Srividya Arjuna1, Gunimala Chakraborty1, Rajesh Venkataram2, Pandyanda Nanjappa Dechamma1, Anirban Chakraborty1
1 Division of Molecular Genetics and Cancer, Nitte University Centre for Science Education and Research, Nitte (Deemed to be University), Mangaluru, Karnataka, India
2 Department of Pulmonary Medicine, K S Hegde Medical Academy, Nitte (Deemed to be University), Mangaluru, Karnataka, India
| Date of Submission | 24-Mar-2020 |
| Date of Decision | 10-Apr-2020 |
| Date of Acceptance | 17-Apr-2020 |
| Date of Web Publication | 11-Jun-2020 |
Correspondence Address:
Anirban Chakraborty
Division of Molecular Genetics and Cancer, Nitte University Centre for Science Education and Research, Nitte (Deemed to be University), Kotekar-Beeri Road, Deralakatte, Mangaluru – 575 018, Karnataka
India
Source of Support: None, Conflict of Interest: None
DOI: 10.4103/jcar.JCar_6_20
Abstract
INTRODUCTION: Targeted therapy using specific inhibitors against tyrosine kinases (TKs) is a paradigm in non-small-cell lung cancer management. However, the success of TK inhibitor (TKI) therapy depends on certain activating or acquired mutations, which render sensitivity or resistance to TKIs in the patients. The acquisition of epidermal growth factor receptor (EGFR) T790M point mutation is the most common mechanism of resistance to TKI in non-small cell lung cancer. A number of molecular strategies are now available for molecular testing of non-small cell lung cancers. However, almost all of them are cost-intensive and laborious and require high-end advanced equipment. Thus, assays that are rapid, simple, and cost-effective, yet sensitive, are most ideal in clinical settings for screening such therapeutically relevant mutations.
MATERIALS AND METHODS: Allele-specific loop-mediated isothermal amplification assay (AS-LAMP), which is a variant of the original LAMP assay, is a promising diagnostic technique for screening single-nucleotide polymorphisms. Using commercially available plasmid constructs as template DNA, AS-LAMP assay for EGFR T790M mutation was optimized with six different sets of reaction mixture containing varying concentrations of buffer and primers. The results of AS-LAMP assay were further validated by ultrasensitive droplet digital polymerase chain reaction.
RESULTS: Only one of the six sets of reaction mixture could accurately distinguish between wild type and mutated DNA, indicating that the primers and buffer are the two most critical components that determine the accuracy of AS-LAMP. The optimized AS-LAMP assay was further used to screen germ line and somatic T790M mutations in non-small cell lung cancer using blood and tissue samples collected from patients.
CONCLUSION: Development of an accurate and rapid diagnostic assay that can detect resistant mutations without the need for sequencing is highly useful for clinicians in deciding on the eligibility of patients for TKI therapy. Considering its several inherent advantages, AS-LAMP assay could become an effective molecular tool for screening baseline or acquired EGFR T790M mutations in non-small cell lung cancer patients.
Keywords: Allele-specific loop-mediated isothermal amplification, genetic mutations, molecular testing, single nucleotide polymorphism, T790M point mutation, targeted therapy
| How to cite this article: Arjuna S, Chakraborty G, Venkataram R, Dechamma PN, Chakraborty A. Detection of epidermal growth factor receptor T790M mutation by allele-specific loop mediated isothermal amplification. J Carcinog 2020;19:3 |
| How to cite this URL: Arjuna S, Chakraborty G, Venkataram R, Dechamma PN, Chakraborty A. Detection of epidermal growth factor receptor T790M mutation by allele-specific loop mediated isothermal amplification. J Carcinog [serial online] 2020 [cited 2021 Oct 15];19:3. Available from: https://carcinogenesis.com/text.asp?2020/19/1/3/286536 |
Introduction
Mutation profiling is crucial in understanding the genetic basis of complex diseases like cancer. Single-nucleotide polymorphisms (SNPs) are important biomarkers, which show great relevance in diagnostics or in therapeutics. Amplification refractory mutation system polymerase chain reaction (PCR), allele-specific PCR (AS-PCR), real-time PCR, and PCR-restriction fragment length polymorphism are commonly usedin vitro diagnostic techniques for screening genomic mutations. However, these recognized molecular techniques are generally cost intensive and often become unaffordable for economically marginalized patients. Loop-mediated isothermal amplification (LAMP) assay is a cost-effective molecular technique, which has seen tremendous applications in the field of infectious disease diagnosis.[1] LAMP amplifies nucleic acids under isothermal conditions using four primers that target six regions in a particular sequence. The method uses Bst polymerase enzyme that possess strand displacement activity. LAMP assays are known for their rapidity, specificity, and sensitivity. Since its discovery in 2000, there have been many improvisations of the basic technique and several variants of LAMP assays have been reported.[2] In recent years, a number of LAMP variants have been developed for screening SNPs.[3] LAMP assay has distinct advantages over conventional amplification techniques such as PCR and other isothermal amplification techniques in terms of high efficiency, rapidity, specificity, and sensitivity.[2],[4] However, false-positive amplification, which is mainly due to the carry over contamination, is one of the major issues in LAMP assays. Indeed, different methods have been adapted to control the false-positive amplifications in LAMP.[5]
AS-LAMP works on the same principle as LAMP with an added advantage of discriminating even a single-nucleotide difference. AS-LAMP assay is being considered as an effective and promising diagnostic technique for screening SNPs.[6] Our focus in this study was to develop an AS-LAMP assay for screening T790M point mutation in exon 20 of epidermal growth factor receptor (EGFR) gene in lung cancer. EGFR mutations are the most important clinically relevant biomarkers in lung cancer. Inactivating EGFR through tyrosine kinase inhibitors (EGFR-TKIs) is a very successful strategy of targeted therapy in lung cancer. The exon 20 of EGFR gene, which is a part of tyrosine kinase (TK) domain, represents one of the most frequently mutated regions conferring resistance to TKIs.[7],[8],[9] Therefore, these mutations form a potential prognostic biomarker as far as the treatment outcome is concerned.[10] Although gene sequencing is considered the gold standard in mutation analysis,in vitro diagnostics (IVDs) rely on other techniques that can indicate the presence or absence of SNPs in patients without the need for sequencing. However, a majority of these IVDs are cost intensive and demand considerable infrastructure. Thus, molecular tools that can identify SNPs at a relatively lesser cost with higher levels of sensitivity and specificity are desirable. Molecular testing in lung cancer usually focuses on identification of mutations that render resistance or sensitivity to TKI drugs in patients, which allows the clinicians to determine the choice of treatment for the patients. T790M mutation in exon 20 of EGFR gene is the most relevant TKI-resistant mutation in non-small cell lung cancer. Here, we report the development of an AS-LAMP assay for screening T790M mutation, which has tremendous potential for routine diagnosis and in point-of-care testing as an alternative cost-effective molecular tool.
Materials and Methods
Ethics
The study was approved by the Central Ethics Committee of Nitte (Deemed to be University), and the experiments were performed using standard ethical practices as per the International Guidelines. The patient samples were collected after obtaining informed consent.
Source of DNA
The optimization of the AS-LAMP assay was carried out using commercially available plasmid constructs. Two EGFR plasmids, one harboring T790M point mutation (pBabeEGFR T790M, #32070) and the other with wild type EGFR sequence (EGFR WT, #11011), were procured from Addgene, a non-profit plasmid repository (https://www.addgene.org/). The plasmids were cloned and the mutation in the target region was confirmed by capillary sequencing.
Primer design
A typical LAMP assay requires a minimum of four primers, two outer (F3 and B3) and two inner (forward inner primer [FIP] and backward inner primer [BIP]). In case of AS-LAMP, the BIP primer is designed separately, one each for wild type and mutated sequence. The modifications are usually made in the 5'end of the BIP to differentiate between the mutant and wild-type DNA. The rest of the primers remain same. Our BIP primer was mutant specific, which allowed amplification only in case of mutant DNA and not in the presence of wild type DNA. The locations of the primers in EGFR gene are provided in [Figure 1].
 |
Figure 1: Primer location in EGFR gene. The nucleotide sequence of exon 20 of human EGFR gene used for designing the primers for this study. The sequence in black indicates the sense strand, whereas the sequence in blue indicates the antisense strand. The primer modification site targeting threonine residue at position 790 is shown in bold and asterisks. NCBI RefSeq: NG_007726.3 Click here to view |
Optimization of loop-mediated isothermal amplification assay parameters
The LAMP assays were performed using a commercially available LAMP kit (Eiken Chemical, Japan). Optimization of parameters such as temperature, time, sensitivity, and specificity was carried out using the wild-type LAMP primers and wild-type plasmid DNA construct ( EGFR WT, #11011). The reaction mixtures for optimization assays were prepared as per the manufacturer&s instructions, which included 12.5μl reaction mixture containing 20 μM each of FIP and BIP and 2.5 μM of F3 and B3.
Temperature and time optimization
This was carried out by performing LAMP at different temperatures and time intervals to obtain maximum yield. The amplification was followed by heat inactivation at high temperature of 80°C for 5 min. The results were analyzed by agarose gel electrophoresis and visualized using Ultra Violet (UV)_ Transilluminator (Gel Doc, BioRad, USA).
Determination of specificity of loop-mediated isothermal amplification assay primers
To determine the specificity of AS-LAMP assay, the addition of primers in the reaction mixture was modulated by gradual elimination of one primer at a time in each set. The reaction was performed at 65°C for 45 min and inactivated at 80°C for 5 min.
Determination of sensitivity of LAMP assay
The sensitivity of AS-LAMP assay was done using tenfold serial dilution of DNA up to nine successive dilutions with the starting concentration of 100ng of total DNA. The reaction was performed at optimum temperature and time. The results were analyzed by gel electrophoresis and visualized through UV Transilluminator (Gel Doc, BioRad, USA).
Optimization of allele-specific loop-mediated isothermal amplification assay components
rimer and buffer concentration
To start with, standard primer and buffer concentration, as recommended by the manufacturer, was used for AS-LAMP. However, non-specific amplifications (wild-type primers showing positive amplification with a mutant plasmid DNA as a template and vice versa) were encountered, which led to further optimization of reaction components. A total of six combinations containing varying concentrations of primers and buffers were randomly chosen ranging from 2 μM to 20 μM. Other parameters were as per the standard protocol. AS-LAMP assays were performed using wild-type/mutant plasmid DNA for each of these sets. The primer–buffer set that was able to differentiate the mutant plasmid DNA from that of wild type plasmid DNA was considered as a positive readout, while those that could not differentiate between the two types of plasmid DNAs were considered as negative readout.
Screening of clinical cases for T790M mutation by allele-specific loop-mediated isothermal amplification assay and validation by droplet digital polymerase chain reaction
The optimized AS-LAMP assay was used to screen T790M mutation in a total of 40 blood samples and 50 tissue samples collected from patients after obtaining the individual informed consents. The AS-LAMP results were further validated by droplet digital PCR (ddPCR, Bio Rad, USA). In ddPCR, tissue and plasmid DNA was used, and the methodology was followed as per the manufacturer's instructions. In ddPCR, the principle of rare mutation detection relies on a single set of primers and two competitive probes: one detecting the wild-type allele (HEX-labeled probe) and another detecting the mutant allele (FAM-labeled probe). The presence of only FAM-labeled droplets would indicate a homozygous mutation, whereas the presence of both FAM and HEX-labeled droplets would indicate a heterozygous mutation. Commercially available T790M Prime PCR DD PCR assay was used (Bio Rad, USA).
Results
Optimization of basic parameters loop-mediated isothermal amplification assay
Since AS-LAMP assay is a variant of original LAMP assay, it was necessary to optimize the basic parameters of LAMP with regard to the temperature, time, specificity, and sensitivity before proceeding with the optimization of AS-LAMP assay. The results of basic parameter optimization are shown in [Figure 2]. The optimum temperature and time chosen were 65°C for 45 min, respectively [Figure 2]a and [Figure 2]b. As for specificity, LAMP reaction was seen only when all the four primers were present and not in assays when any of the four primers were absent [Figure 2]c. With regard to the minimum amount of DNA necessary for LAMP assay, the amplification was observed even at minimal concentration of 10-7 dilution, which was equivalent to 10 fg of DNA [Figure 2]d.
| Figure 2: Gel images of optimization conditions for LAMP assay. (a) Temperature optimization: Lanes marked as 55, 60, 62, 65, and 70 indicate thedifferent incubation temperatures (°C), NC: Negative control. (b) Time optimization: Lanes marked as 30, 45, 60, and 75 indicate the incubation time (in min), NC: Negative control. (c) Specificity of LAMP assay: Results of LAMP assay on addition of four primers (L4), three primers (L3), two primers (L2), and one primer (L1), NC: Negative control. (d) Sensitivity of LAMP assay: 100-10-9 indicate the tenfold serial dilution with starting concentration of 100 ng (100) of DNA, NC: Negative control Click here to view |
Determination of optimum primer and buffer concentration for allele-specific loop mediated isothermal amplification assay
The determination of optimum primer and buffer concentration was based on the criterion that the chosen primer set should be able to differentiate the mutant DNA from wild-type DNA. In the standardization experiments, the chosen primer set was mutant specific, which was expected to show a positive reaction only in the presence of mutant plasmid DNA and not with wild-type plasmid DNA. Of the six different reaction cocktails with each one containing different concentrations of primers and buffer with rest of the reaction component unchanged, only one set (Set 6) could distinguish the mutant DNA from wild-type DNA. The rest of the five sets either failed to amplify or amplified both the mutant- and the wild-type DNA [Figure 3]. This reaction cocktail was repeated several times to confirm the specificity and the reproducibility of the assay, and consistent results were obtained in all the attempts.
| Figure 3: Gel image of six sets of reaction mixtures used for optimization of AS-LAMP assay: Set 1 to Set 6 indicate the six different combinations of reaction cocktail containing variable concentrations of primer and buffer. DNA template included plasmid DNA containing EGFR gene with T790M mutation (1) and plasmid containing wild type EGFR gene (2). Lane 3 indicates the negative control Click here to view |
Screening of clinical samples by allele-specific loop-mediated isothermal amplification assay
To determine the feasibility of the standardized assay in screening patients' samples for target mutation (T790M), we extracted the total DNA from 40 blood samples and 50 tissue samples obtained from lung cancer cases. The extracted DNA was used as a template for AS-LAMP assay to screen for germ line or somatic T790M point mutation. The screening revealed that only 1 of 50 tissue samples was positive for T790M mutation. None of the blood samples were positive for T790M mutation. Among the 40 blood samples, 34 (85%) were of non-small cell lung cancer type and 3 (7.5%) were small cell lung cancer. The histological classification of the remaining 3 (7.5%) was not clear. With regard to the gender, 37 of 40 (92.5%) cases were male and 3 (7.5%) were female. Twenty-one of forty (52.5%) patients were smokers, whereas 10 (25%) were nonsmokers. In 9 cases (22.5%), the smoking status was not known.
In case of tissue samples, 39 of 50 (78%) were of non-small cell lung cancer type and 5 (10%) belonged to small cell lung cancer. In the remaining 6 (12%) cases, the histological classification was inconclusive. Of the 50 samples analyzed, 44 (88%) were male and 6 (12%) were female. Thirty-one cases (62%) were smokers, whereas 15 (30%) were nonsmokers. In 4 cases (8%), the smoking status was not available.
Validation of allele-specific loop-mediated isothermal amplification assay results by droplet digital polymerase chain reaction
To further validate the results of AS-LAMP assay, ddPCR for T790M mutation was performed using the DNA of the tissue sample that showed positive by AS-LAMP. ddPCR is the third generation ultrasensitive endpoint PCR that utilizes water–oil emulsion droplets for amplification and provides absolute quantification of target DNA with high levels of accuracy and precision. It relies on partitioning effect where each droplet is subject to standard endpoint PCR. The results of ddPCR were consistent with those obtained from AS-LAMP assay. As shown in [Figure 4], the tissue sample (lane D04), which was positive for T790M mutation by AS-LAMP, showed a significant number of blue (FAM-labeled) droplets indicating the presence of mutant DNA. Expectedly, the sample was positive for the wild-type allele for T790M as indicated by the presence of green droplets (HEX-labeled) in the lower panel of [Figure 4], suggesting the presence of heterozygous mutation. The no template control (H04) showed negative for both FAM-labeled and HEX-labeled droplets indicating the correctness of the assay.
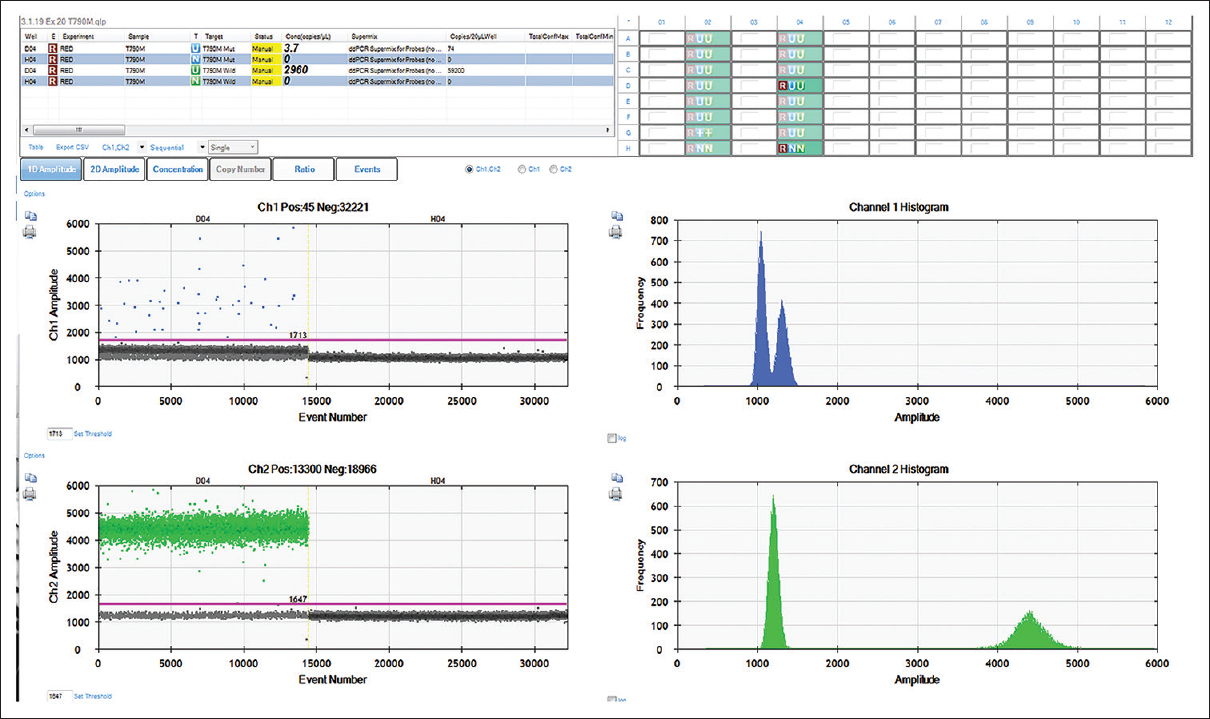 |
Figure 4: A representative image of EGFR T790M mutation assay by ddPCR: A representative image (1D plot from QuantaLife software) of EGFR T790M mutation assay by ddPCR. Two probes, one labeled with FAM (for mutant) and another with HEX (wild type), were used. The blue dots indicate the presence of FAM-labeled droplets positive for mutant allele (D04, upper panel), whereas green dots indicate the HEX-labeled droplets positive for wild-type allele (D04, lower panel). The channel amplitudes are plotted in Y-axis separately FAM and HEX, whereas the events (number of droplets) are plotted in X-axis. The magenta line indicates the baseline and the black dots indicate droplets without any DNA Click here to view |
Discussion
Conventionally, gene mutations are identified by directly sequencing the gene of interest. However, in recent years, several novel methods have been developed that can effectively identify the presence or absence of mutation without sequencing. LAMP, which was developed almost a decade ago, is now accepted as a cost-effective alternative to PCR-based detection techniques.[11] Indeed, LAMP and its several other variants offer rapid, accurate, and cost-effective diagnosis of infectious diseases.[12] It is now being appreciated as an alternative tool for SNP identification.[3] AS-LAMP has been developed for detecting activating mutations in JAK2 gene in chronic myeloproliferative neoplasms [13] and for point mutations that contribute to drug resistance in fungi and in mosquitoes.[14],[15] In this study, an AS-LAMP assay has been developed for detecting T790M point mutation in exon 20 of EGFR gene. The primers for AS-LAMP were designed in such a way that it amplified only the mutated DNA but not the wild-type DNA. Surprisingly, the standard primer and buffer concentration as recommended by the manufacturer could not distinguish between the mutant- and the wild-type DNA. This prompted the standardization with varying concentrations of primers and buffer in the reaction mixture. The results of this study clearly demonstrate that the concentration of the buffer and the primers must be standardized when LAMP is used for discriminating a single-nucleotide difference. The applicability of the developed assay as a potential IVD technique was further demonstrated by screening a large number of patient samples for the target mutation and re-validation using droplet digital PCR. Since the standardization of the assay was done using commercially available plasmid constructs carrying the desired mutation, it was necessary to show the reproducibility of the results obtained in AS-LAMP by other techniques Therefore, ddPCR was performed using the plasmid DNAs, both wild-type and mutant, with varying concentrations of template DNA. As shown in [Figure 5], the wild-type plasmid DNA (lanes B02 to D02) showed only HEX-labeled droplets (green) in the lower panel as expected for the wild-type probe, whereas the mutant plasmid DNA (lanes E02 to G02) showed a very high number of FAM-labeled droplets (blue) as expected for the probe that binds to mutant allele. These lanes (E02 to G02) did show a few green droplets, which might be due to the fact that the mutant plasmid was created by site-directed mutagenesis strategy and it might have resulted in a heterozygous condition. Nevertheless, the number of blue droplets was much higher in these lanes suggesting the clear presence of mutant allele. As expected, the no template control (lane A02) did not show any FAM or HEX-labeled droplets indicating the accuracy of the assay.
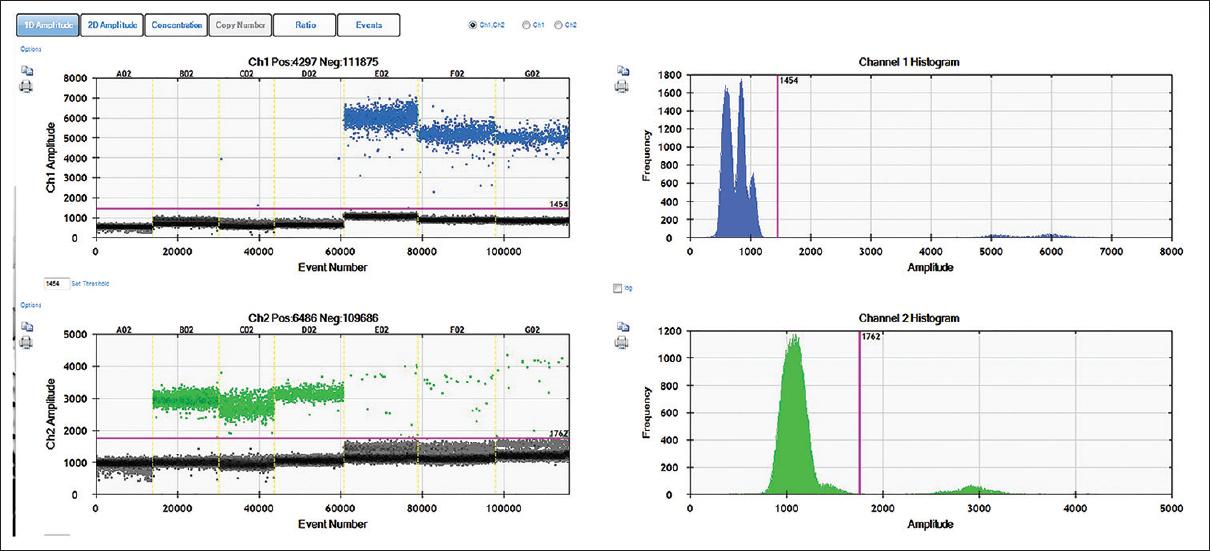 |
Figure 5: Validation of wild type and mutant EGFR plasmid DNA by ddPCR: The 1D plot of QuantLife software showing the results of EGFR T790M mutation assay by ddPCR using wild-type and mutant plasmids as DNA templates. Two probes, one labeled with FAM (for mutant) and another with HEX (wild type), were used. Lanes B02 to D02 contained EGFR WT, #11011 plasmid DNA with varying concentration (20,000 copies, 10,000 copies and 5,000 copies respectively), whereas lanes E02 to G02 contained pBabeEGFR T790M, #32070 plasmid DNA varying concentration (20,000 copies, 10,000 copies, and 5000 copies respectively). The blue dots indicate the presence of FAM-labeled droplets positive for mutant allele (E02 to G02, upper panel), whereas green dots indicate the HEX-labeled droplets positive for wild-type allele (B02 to D02, lower panel). The channel amplitudes are plotted in Y-axis separately for FAM and HEX, whereas the events (number of droplets) are plotted in X-axis. The magenta line indicates the baseline and the black dots indicate droplets without any DNA Click here to view |
Of the ninety samples screened, only one tissue sample showed positive for T790M mutation. This was not surprising given the fact that germ line T790M mutation is extremely rare and is estimated to occur in 1% of non-small cell lung cancer patients.[16],[17] The presence of germ line EGFR T790M mutations is believed to be associated with inherited susceptibility to non-small cell lung cancers and is known to confer a carrier status to the patients.[16] The somatic mutations are rarely seen prior to treatment and are generally acquired in patients undergoing TKI therapy. Acquisition of EGFR T790M mutation is the most common mechanism of resistance to TKI therapy, which occurs in 60% of the patients.[9] However, primary de novo EGFR T790M somatic mutations have been reported in patients prior to TKI treatment, who carry dual or multiple EGFR mutations.[18] In our study, we have observed that 1 of 50 tissue samples (2%) showed the presence of primary de novo T790M mutation. A recent study by Li et al.[19] reported that the primary T790M mutation occurred in 0.5% of TKI-naive patients, whereas in TKI-relapsed patients, it was as high as 49.7%.
We have confirmed our data with the ultrasensitive ddPCR, which is capable of picking up rare mutations that are otherwise missed by standard Sanger sequencing. Although the sensitivity of a genotyping tool varies depending on the method used, we believe that the sensitivity of AS-LAMP method developed here is comparable to other PCR-based techniques.
To summarize, this study describes the development of an AS-LAMP assays for the detection of EGFR T790M mutation in non-small cell lung cancer cases. Recently, a novel LAMP method that employed a fluorophore-labeled probe, in addition to the LAMP primers, was used to detect EGFR sensitizing mutations in surgically resected tumor tissues from patients with pulmonary adenocarcinoma and its efficiency was compared with Therascreen qPCR assay.[20] Although the results were consistent in both the methods, the authors did not include EGFR T790M mutation, which is associated with resistance to TKI-therapy. In addition, their analysis was not focused on determining germ line status or primary de novo T790M mutation in the tissues. Nevertheless, the article supports the use of LAMP as a cost-effective alternative for screening EGFR mutations in lung cancer.
In our study, the lack of positive cases in blood samples indicated that germ line mutation of EGFR is extremely rare and it is the somatic gain of function mutation in EGFR that contribute to the resistance to TKI therapy. However, even with tissue samples, only one positive case was encountered, which was most likely because of the higher incidence of activating EGFR mutations such as L858R and Exon 19 deletion in Asians and less incidence of upfront or baseline EGFR T790M point mutation in TKI-naive patients. The results of this study clearly demonstrate the feasibility of AS-LAMP in screening patients for EGFR T790M mutation. The assay could be extended to tumor-derived circulating DNAs and could provide alternative options for liquid biopsy techniques, which are currently highly cost intensive.[21]
Conclusion
AS-LAMP assays have tremendous potential for routine diagnosis and in point-of-care testing. The test developed here is cost-effective and does not require sophisticated equipment. It can be effectively utilized for prognostic application in lung cancer management in a non-invasive manner, particularly for detecting the emergence of this resistance mutation in lung cancer patients undergoing TKI therapy using cell-free DNA. The assay could also be used for screening germ line T790M mutations, which is a predisposing factor for never smokers to lung cancer.
Acknowledgments
The support from Nitte (Deemed to the University) in the form of a PhD fellowship to thefirst author and the financial support provided through the intramural research grant (NUFR3/2016/05-04) to the corresponding author are gratefully acknowledged.
Financial support and sponsorship
Intramural research grant (NUFR3/2016/05-04) was awarded to the corresponding author.
Conflicts of interest
There are no conflicts of interest
References
| 1. | |
| 2. | |
| 3. | |
| 4. | |
| 5. | |
| 6. | |
| 7. | |
| 8. | |
| 9. | |
| 10. | |
| 11. | |
| 12. | |
| 13. | |
| 14. | |
| 15. | |
| 16. | |
| 17. | |
| 18. | |
| 19. | |
| 20. | |
| 21. |

