Shashi Ranjan Mani Yadav1, Bela Goyal1, Raman Kumar1, Sweety Gupta2, Amit Gupta3, Anissa Atif Mirza1, Gaurav Sharma2, Shalinee Rao4, Rajesh Pasricha2, Manoj Gupta2
1 Department of Biochemistry, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India
2 Department of Radiation Oncology, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India
3 Department of Surgery, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India
4 Department of Pathology and Laboratory Medicine, All India Institute of Medical Sciences, Rishikesh, Uttarakhand, India
| Date of Submission | 26-Jul-2020 |
| Date of Decision | 01-Sep-2020 |
| Date of Acceptance | 12-Sep-2020 |
| Date of Web Publication | 20-Nov-2020 |
Correspondence Address:
Sweety Gupta
Department of Radiation Oncology, All India Institute of Medical Sciences, Rishikesh – 249 203, Uttarakhand
India
Source of Support: None, Conflict of Interest: None
DOI: 10.4103/jcar.JCar_18_20
Abstract
INTRODUCTION: Lung cancer (LC), among all other cancers, is the leading cause of death worldwide, while the third most common cancer-causing mortality in India. Several techniques of the assay for early detection of cancer that improve survival rates have been employed in tissues and cell lines. Reverse transcriptase quantitative polymerase chain reaction (RTqPCR) is one of the most common techniques employed for gene expression studies for the normalization of a target gene using a reference gene (RG). The present study used the three most common RGs: glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-Actin, and 18s ribosomal ribonucleic acid (18s rRNA), which were assessed by qPCR to validate, as of which is a more effective RG in blood samples of LC patients.
MATERIALS AND METHODS: A total of thirty participants with LC of non-small cell and small cell type were included along with twenty healthy controls. Ribonucleic acid (RNA) isolated from peripheral blood mononuclear cells was quantified, prepared for complementary deoxyribose nucleic acid synthesis, and analyzed for expression of three RG on RTqPCR.
RESULTS: Expression levels as Ct values of studied RG were reported as mean ± standard deviation for GAPDH (26.97 ± 5.107), β-actin (20.5 ± 2.3), and 18s rRNA (25.10 ± 4.075). GAPDH showed the lowest expression, whereas β-actin showed the highest expression among the studied RG in subjects of LC. The expression of GAPDH and 18s rRNA were statistically significantly lower than β-actin (p < 0.0001), whereas expression levels of GAPDH and 18s rRNA were comparable. However, the expression level of only β-actin in LC patients was comparable with healthy controls with P < 0.1611 at 95% confidence interval.
CONCLUSION: It is concluded that β -actin may be considered the most suitable RG isolated and studied from peripheral blood mononuclear cells using RT qPCR in LC.
Keywords: Circulating tumor cells, liquid biopsy, lung cancer, reference genes
| How to cite this article: Yadav SR, Goyal B, Kumar R, Gupta S, Gupta A, Mirza AA, Sharma G, Rao S, Pasricha R, Gupta M. Identification of suitable reference genes in blood samples of carcinoma lung patients using quantitative real-time polymerase chain reaction. J Carcinog 2020;19:11 |
| How to cite this URL: Yadav SR, Goyal B, Kumar R, Gupta S, Gupta A, Mirza AA, Sharma G, Rao S, Pasricha R, Gupta M. Identification of suitable reference genes in blood samples of carcinoma lung patients using quantitative real-time polymerase chain reaction. J Carcinog [serial online] 2020 [cited 2021 Oct 13];19:11. Available from: https://carcinogenesis.com/text.asp?2020/19/1/11/300977 |
Shashi Ranjan Mani Yadav and Bela Goyal contributed equally towards manuscript
Introduction
Lung cancer (LC), among all other cancer, is the leading cause of death among populations of the world,[1] whereas it is the third most common leading cause of cancer-related mortality in India after breast and cervical cancer.[2] Survival rates of cancer are entirely dependent on their earlier detection. Currently, diagnosis is made by clinical examination for chest findings, any peripheral lymphadenopathy, radiological imaging chest x-ray, contrast-enhanced computed tomography CECT scan, and histopathology, or biopsy. The tissue collection process is an invasive procedure and limited by sensitivity and specificity. Immunohistochemistry and molecular testing are being developed for the identification of subtype and tissue of origin as the lung. Circulating tumour cells (CTCs) were initially detected in 1869 by Thomas Ashworth in the blood sample of a breast cancer patient. They are shed from primary tumor masses and deposit at other sites through the bloodstream. They have been detected in most epithelial cancers, which include either breast, prostate, colon, and lung. CTC analyses are considered a real-time ”liquid biopsy” for patients with cancer.[3],[4],[5],[6],[7]
Real-Time reverse transcriptase quantitative Polymerase Chain Reaction (RT-qPCR) technology has been the most commonly employed technique in the early diagnosis and detection of LC based on quantifying the expression of ribonucleic acid (RNA) transcript. The qPCR data in genomic studies must be normalized with an appropriate internal standard, the so-called reference gene (RG), where the outcomes are expressed as ”target to reference ratio”.” RGs are actively expressed to carry out normal cellular activities and prevail a similar degree of expression across all cells and tissues in varying experimental conditions.[1],[8],[9] Some reports have already validated one or more RG for RT-qPCR in different cancers.[1],[10],[11],[12],[13],[14] While there is the inconsistency of results of RG in tissue and cell lines, several recent studies have employed either of these various endogenous control genes, such as Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, 18s ribosomal RNA (18s rRNA), TATA-binding protein, phenylalanine hydroxylase[1] ESD, BTF3, HIST1H2BC, RPL30, and YAP1[8] for qPCR. Still, none has been evaluated in blood samples of LC patients. This is important as the liquid biopsy is emerging as the mainstay of detecting molecular markers in LC. In the present study, three of the most common RGs: GAPDH, β-actin and 18S rRNA were assessed by qPCR to validate, as of which is the more effective RG in blood samples of LC. Therefore the current study was planned to identify suitable RG in blood samples of LC patients and healthy controls. This study may provide insight to promote a better understanding of the RG expression in LC and also will have remarkable application in establishing the normalization of levels of target gene expression in the prospects of cancer research.
Materials and Methods
Thirty LC patients and twenty healthy controls (HCs) were enrolled in the study after obtaining written informed consent. The study was approved by the institutional ethical committee (83/IEC/EM/2016). Informed consent was obtained from all patients and healthy controls. The blood sample was collected from each participant through venipuncture in ethylene diamine tetraacetic acid vial after discarding the first 2ml to avoid epithelial cell contamination and mixed well. Blood sample was immediately processed for peripheral blood mononuclear cells (PBMCs) isolation.
Peripheral blood mononuclear cells isolation
The 3ml of whole blood was pipetted in a Falcon tube, adding an equal proportion of phosphate-buffered saline (PBS; pH 8.2) buffer and mixed well. The mixture is then layered on the Histopaque reagents (Sigma-Aldrich) and centrifuged at 1300 rpm for 20 minutes in a break- free centrifuge. The second thick PBMCs layer to the upper layer of the four-layer series is collected and transferred into diethyl pyro-carbonate treated sterile Eppendorf tube. The PBMCs collected are washed with PBS buffer with brief centrifugation and then stored for further use.
Ribonucleic acid isolation, purification, and complementary deoxyribose nucleic acid preparation
Total RNA was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA) as described previously.[15]
The isolated RNA was purified to remove any genomic Deoxyribose nucleic acid (DNA) contamination by treating with DNase treatment (Qiagen, USA) according to the manufacturer’s instructions. Total RNA concentration was measured on the TECAN NanoQuant plate (TECAN Infinite 200 PRO, Switzerland) at OD260nm, and the purity was confirmed with the ratio measured at 260/280.
The first strand complementary DNA (cDNA) was synthesized using the iScript cDNA synthesis kit (Bio-Rad). The concentration of total RNA for cDNA synthesis was kept at 500ng and 4μl of 5x iScript Reaction Mix added with 1μl of iScript reverse transcriptase (RT). The total volume of reaction was made up to 20μl by nuclease-free water. The reaction protocol for reaction mix follows with incubation for priming at 25° for 5 min and reverse transcription at 46° for 20 min following RT inactivation at 95° for 1 min.
Reverse transcriptase-quantitative polymerase chain reaction
Three endogenous control genes (GAPDH, β-actin, and 18s rRNA) were measured by qPCR using the iTaq UniverSYBR Green SMX (Bio-Rad) Bio-Rad RT-PCR system. Specific primers of genes were designed and developed utilizing Primer3 Input software version 0.4.0 and purchased from IDT biotechnologies. The sequences designed for the primers are mentioned in [Table 1].
| Table 1: Primer sequences for reference genes Click here to view |
The final volume of PCR reactions was prepared in 20μL with 2x SYBR Green Master Mix 5μl, forward primer 0.5μl, reverse primer 0.5μl and cDNA 1μl, and the total volume was made up to 20μl with nuclease-free water.
The thermal cycling condition comprised an initial incubation at 95° for 5 min, followed by 34 cycles of denaturation at 95° for 30s, and annealing at 56° for 30s and extension at 70° for 30s. The final extension was promoted at 70° for 10 min. Each measurement was performed in duplicate along with no template as control, and the threshold cycle (Ct) was determined.
Statistical analysis
GraphPad Prism software version 5.0 was used to analyze the results. Paired t-test was used to compare Ct values of RGs among LC. Unpaired student t-test was used to compare Ct values of RGs between LC and HCs. P < 0.05 was considered to be statistically significant.
Results
Thirty patients of diagnosed cases of non-small cell LC (NSCLC)(86.66%) and small cell LC (SCLC) (13.33%) were included and underwent complete staging workup and tumor evaluation. According to the criteria of the American Joint Commission on Cancer 8th Edition (AJCC), clinical stages were determined, and the pathological type was diagnosed as lung adenocarcinoma, squamous cell carcinoma, and small cell carcinoma. Patients with synchronous or metachronous second malignancy or treated for carcinoma lung were excluded from the study. In addition, twenty 29-52 years (mean – 38.5 years) healthy volunteers were selected as the control group. Patient and healthy control characteristics are given in [Table 2].
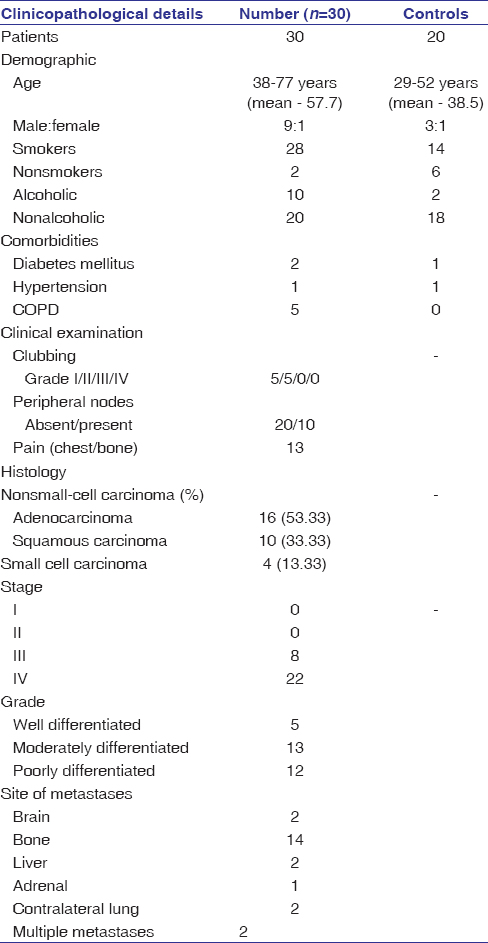 |
Table 2: Patients and healthy control characteristics Click here to view |
We determined levels of three different RGs, namely GAPDH, β-actin, and 18s rRNA, with qPCR in participants included. Expression levels based on mean ± standard deviation Ct values of three RGs in LCs were GAPDH (26.97 ± 5.107), β-actin (20.5 ± 2.3) and 18s rRNA (25.10 ± 4.075) and in HCs were (21.08 ± 1.806), (19.77 ± 2.324) and (19.70 ± 2.920) respectively.
Differential expression levels of the RGs among LCs were observed, as represented in [Table 3]. GAPDH showed the lowest expression, whereas β-actin showed the highest expression among the studied RGs in LCs. The expression of GAPDH and 18s rRNA was significantly lower than β-actin.
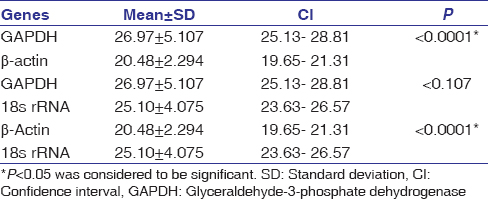 |
Table 3: Expressional behavior of three reference genes among lung cancers Click here to view |
When the expression of RGs was compared between LCs and HCs, only β-actin showed comparable expression between LCs and HCs. The expression levels of GAPDH and 18s rRNA in LCs were significantly different from those of HCs with P < 0.001 and P < 0.0001 at 95% confidence interval (CI) respectively. While the expression level of β-actin in LCs was comparable with HCs with P < 0.1611 at 95% CI, as shown in [Table 4] and [Figure 1].
| Table 4: Expression of reference genes in lung cancer patients and healthy controls Click here to view |
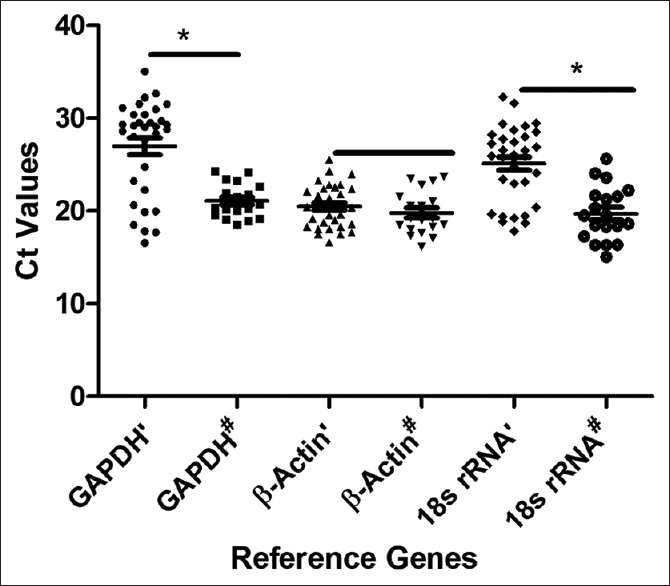 |
Figure 1: Differential expression of Lung Cancer versus Healthy Controls: The difference in expression of GAPDH’ is significant from GAPDH#; β-actin’ is relatively expressed similar to β-Actin#; 18s rRNA’ expression is significantly different from 18s rRNA#; while β-actin’ is significantly different from GAPDH’ and 18s rRNA’ and GAPDH#, β-Actin# and 18s rRNA are relatively similar in expression. GAPDH’: GAPDH in LC, GAPDH#: GAPDH in HC, β-Actin’: β-Actin in LC, β-Actin#: β-Actin in HC, 18s rRNA’: 18s rRNA in LC, 18s rRNA#: 18s rRNA in HC. *P < 0.05: Significant differences Click here to view |
Discussion
As the research has advanced to the molecular level involving RNA transcript in establishing diagnosis and prognosis of the disease, the normalization of the genes with suitable RGsto determine the expression of RNA is of utmost importance. The ideal RGs should behave similarly, unaffected in all cells and tissues irrespective of diseases and experimental conditions. Many RGs have been documented over the years in tissue biopsy or cell lines, but we studied three of the most common and established/susceptible RGs GAPDH, β-actin, and 18s rRNA in blood samples of LC (NSCLC and SCLC) and also compared them with HC. β-actin as cytoskeletal protein maintains the cellular shape harboring fibronectin with transmembrane glycoproteins in the extracellular matrix with actin microfilaments residing in the cytoplasmic side of the membrane. This need to maintain stability in cellular shape causes controlled regulation of β-actin expression.[16]
GAPDH acts as a metabolic enzyme in glycolysis. The reaction catalyzed is allosterically regulated by the consumption of NAD+, which is contributed either by the anaerobic pathway of glycolysis or through the electron transport chain. Consequently, it is evident that GAPDH is expressed equally in all tissues and is tightly regulated. Despite being tightly regulated, GAPDH has been affected by several factors, including evidence of higher expression in tissues with higher energy demands.[1],[17],[18]
18S rRNA is highly recommended as housekeeping genes in several mRNA quantification experiments because of its importance. 18S are ribosomal RNA and are not affected by mitogens, which controls the expression of metabolic enzymes used as RGs. However, 18S have their drawbacks as RGs because they are highly expressed as representing 80% of total cellular RNA and are ribosomal RNA where they miss out on any significant changes in levels of mRNA.[8],[19],[20]
In the present study, when we compared the most commonly used RGs for LCs, GAPDH and 18s rRNA showed significantly higher Ct values with low expression and higher variability. However, when we compared the individual RG gene between LCs and HCs, only β-actin was comparable. In contrast, both GAPDH and 18S rRNA showed significantly decreased expression in LCs as compared to HCs. This corroborates with a previous study where GAPDH exhibited lower expression and higher variability.[1] However, it is contrary to the study by Barber et al., which showed GAPDH to be the ideal RG.[17]
In our study, GAPDH showed low and variable expression. This is in agreement with a previous study by Silviaet al. where they have done a comparison of five putative and seven commercial RGs in LC and did meta-analysis in LC transcriptomes. They identified GAPDH to be one of the most variable RG with low expression in 18 NSCLC patients similar to our study.[18] The plausible explanation is modulation of GAPDH expression due to factors such as hypoxia, insulin, mitogen, and epidermal growth factor – in cancers including LC, resulting in variation in cancer tissue as compared to normal tissue. Moreover, they have hypothesised that acetylated histones may result in the downregulation of GAPDH in cancers by unclear mechanism. Moreover, in a study on LC, YEATS2, a gene that increases the expression of histone acetylases was found to be overexpressed. It was shown that YEATS 2 recruits ATAC complex to chromatin on binding to histone acetylase and results in euchromatin formation and increased proliferation and invasion in lung cancer.[19] Taken together, this implies the role of histone acetylase in reduced expression of GAPDH in LC which could be a plausible explanation for low and variable expression of GAPDH in our samples.
Moreover, GAPDH earlier identified as glycolytic enzymes and therefore, a housekeeping gene has now been demonstrated to be involved in a multitude of other functions such as membrane transport, transcriptional regulation, and itself is dysregulated in cancer.[20] Moreover, studies have shown metabolic remodeling in LC where cancer cells are shown to exhibit differential glycolysis rates and mitochondrial capacities under influence of tissue microenvironment.[21] This may be another reason for the dysregulated expression of GAPDH in LC.
Similarly, in a previous study, 18S rRNA was found to be less stable RG amongst ten RG when analyzed by NormFinder and geNorm software whereas stable with Bestkeeper software in LC.[22] The plausible explanation could be due to the aberrant methylation of promoter regions for genes of 18SrRNA in LC that may result in reduced and variable expression of 18SrRNA.[23] Therefore, epigenetic changes such as acetylation and methylation may be the reason for the observed variable and aberrant expression of otherwise housekeeping genes GAPDH and 18S rRNA in LC patients and needs to be explored in further studies. It has also been studied by Ali et al., where they concluded the same during their study through different statistical program.[22] Therefore, we propose β-Actin to be a suitable RG in blood for non-small cell and small cell LC.
Conclusion
Out of the three most commonly used RG for LC, we found β-Actin to be the most suitable RG for NSCLC and SCLC with comparable expression in LCs and HCs.
Acknowledgment
This study has been supported by the Department of Science and Technology New Delhi under WOS-A Project (SR/WOS-A/LS-605/2016(G).
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
| 1. | |
| 2. | |
| 3. | |
| 4. | |
| 5. | |
| 6. | |
| 7. | |
| 8. | |
| 9. | |
| 10. |
Beillard E, Pallisgaard N, van der Velden VHJ, Bi W, Dee R,et al., Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ”real-time” quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)-A Europe against cancer program. Leukemia 2003;17: 2474–86.
 |
| 11. | |
| 12. | |
| 13. | |
| 14. | |
| 15. | |
| 16. | |
| 17. | |
| 18. | |
| 19. | |
| 20. | |
| 21. | |
| 22. | |
| 23. |
