Spoorti Kulkarni, Monica Solomon, Chetana Chandrashekar, Nisha Shetty, Sunitha Carnelio
Department of Oral and Maxillofacial Pathology, Manipal College of Dental Sciences, Manipal Academy of Higher Education, Manipal, Karnataka, India
| Date of Submission | 29-Apr-2020 |
| Date of Decision | 17-May-2020 |
| Date of Acceptance | 17-Jun-2020 |
| Date of Web Publication | 25-Nov-2020 |
Correspondence Address:
Sunitha Carnelio
Department of Oral and Maxillofacial Pathology, Manipal College of Dental Sciences, Manipal Academy of Higher Education, Manipal – 576 104, Karnataka
India
Source of Support: None, Conflict of Interest: None
DOI: 10.4103/jcar.JCar_13_20
Abstract
BACKGROUND: Spalt-like transcription factor 4 (SALL4) is a stem cell marker that plays a critical role in maintaining the pluripotency and self-renewal of embryonic and hematopoietic stem cells. Only a few studies have been done to apprehend the expression of SALL4 in the potentially malignant oral lesion (leukoplakia with dysplasia) and oral squamous cell carcinoma (OSCC).
AIM: The aim of this study is to evaluate the expression of SALL4 in leukoplakia with dysplasia and OSCC and to correlate the expression of the marker (SALL4) with the various clinicopathological parameters and patient outcome.
MATERIALS AND METHODS: Immunohistochemistry for SALL4 protein was performed on 140 cases: those histopathologically confirmed cases of leukoplakia with dysplasia (n = 30) and OSCC (n = 110). Ten cases of nonepithelial neoplasm (fibroepithelial hyperplasia and excised tissue surrounding impacted third molars) were taken as control. Statistical analyses were applied to evaluate correlations between SALL4 overexpression and clinicopathological features of leukoplakia and OSCC. Survival rates were analyzed using Kaplan–Meier method.
RESULTS: SALL4 positivity was observed to be higher (P = 0.001) in the tumor cells of OSCC with Immuno Reactive Score (IRS) ranging from 0 to 9. Poorly differentiated squamous cell carcinoma (SCC) had paramount higher expression with a median IRS of 6. Similar IRS and above (IRS, 6–9) was observed in Stage I (five cases), which recurred and well-differentiated cases with metastasis (four cases) while in leukoplakia with dysplasia the SALL4 expression was weak with a range of 2–4.
CONCLUSIONS: SALL4 being one of the cancer stem cell molecules plays an important role in the progression of oral cancer, which was evident in this study. This could also account for aggressive clinical behavior. Follow-up of these patients would relate this molecule could be responsible for cancer relapse. Patients diagnosed to have oral epithelial dysplasia had a low expression of SALL4, are under follow-up, although seven cases did transform to SCC. Thus, we conclude, SALL4 may be of prognostic relevance, but in oral epithelial dysplasia, it requires further investigations.
Keywords: Carcinogenesis, chemoradiation, epithelial dysplasia, spalt-like transcription factor 4, squamous cell carcinoma, surgery
| How to cite this article: Kulkarni S, Solomon M, Chandrashekar C, Shetty N, Carnelio S. Spalt-like transcription factor 4 expression in oral epithelial dysplasia and oral squamous cell carcinoma: An immunohistochemical appraisal. J Carcinog 2020;19:12 |
| How to cite this URL: Kulkarni S, Solomon M, Chandrashekar C, Shetty N, Carnelio S. Spalt-like transcription factor 4 expression in oral epithelial dysplasia and oral squamous cell carcinoma: An immunohistochemical appraisal. J Carcinog [serial online] 2020 [cited 2021 Oct 13];19:12. Available from: https://carcinogenesis.com/text.asp?2020/19/1/12/301569 |
Introduction
One of the leading cancers in most of the Asian countries with global distribution is oral squamous cell carcinoma (OSCC), ranking sixth-most common neoplasm in the world. Oral cancer (OC) grounds severe morbidity and mortality rates due to a lack of awareness in the population about the lesion at an early stage. In the present scenario, though the diagnostic and therapeutic interventions in OSCC have improved, the 5-year survival rate continues to remain at about 50%.[1],[2],[3] Thus identifying the molecular marker which can help in early detection of this lethal disease is inevitable and the need of the hour. Recent studies have indicated that cancer stem cells (CSCs: tumor cells with the capability of instigating and sustaining tumor growth) as one of the causes for OC. These CSCs share many molecular similarities to embryonic and normal adult stem cells and have the ability of self-renewal and multi-differentiation.[4],[5],[6]
Spalt-like transcription factor 4 (SALL4) is one of the stemness markers (embryonic and CSC) belonging to zinc finger family that plays an important role in maintaining the pluripotency and self-renewal of embryonic and hematopoietic stem cells.[7],[8] Overexpression of SALL4 has been studied in various human cancers: breast, endometrial, cervical cancer and has been linked to poor prognosis.[9],[10],[11] However, very few studies are noted on head and neck cancer.[12],[13],[14],[15] In the present study, we analyzed the expression of this biomarker (SALL4) in OSCC and leukoplakia with dysplasia. The association between the SALL4 and clinicopathologic parameters was also analyzed and attempted to evaluate the role of this molecular marker in the diagnosis and prognosis of the disease process has been looked at.
Materials and Methods
Formalin-fixed paraffin-embedded tumor blocks (n = 140) of histopathologically diagnosed cases of moderate dysplasia (15) and severe dysplasia (15) that were clinically diagnosed as leucoplakia; OSCC (110) that comprised of well-differentiated (n = 40), moderately differentiated (n = 40) and poorly differentiated SCC (n = 30) were retrieved from the archives of the department. Carcinomas were evaluated using the American Joint Committee on Cancer staging system (eighth edition).[16] The patients’ medical records were accessed to obtain the details about the clinicopathological features of the cases after obtaining ethical approval [Table 1]. Ten cases of nonepithelial neoplasm (fibroepithelial hyperplasia and excised tissue surrounding impacted third molars) were taken as a control for the study. Specimens obtained for the study were between 2014 and 2016, after randomization, and the follow-up period ranged from 3½ to 5 years.
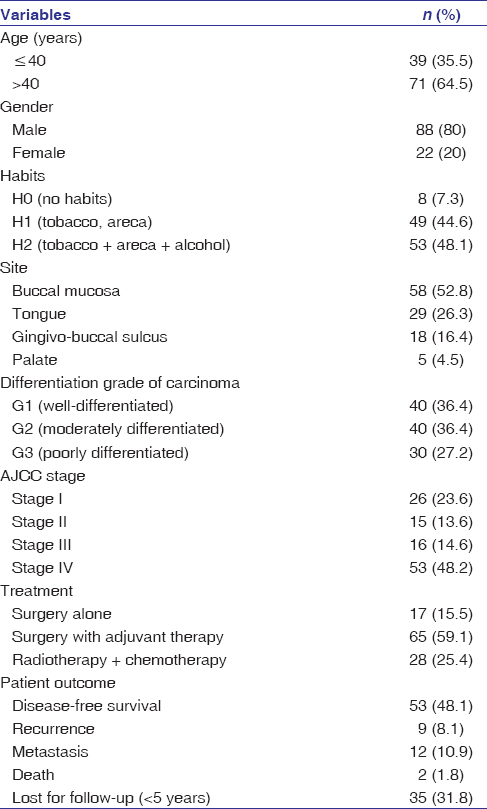 |
Table 1: Clinicopathological parameters of patient with oral squamous cell carcinoma (n=110) Click here to view |
Inclusion criteria
Primary cases of OSCC underwent surgery, while that for potentially malignant lesion included histopathologically proven dysplasia with a history of tobacco use.[17],[18]
Exclusion criteria
Tumors other than OSCC and recurrent OSCC, while that for potentially malignant lesion, excludes other definable lesions with a probable history of tobacco use.
Immunohistochemistry
Four microns thickness formalin-fixed paraffin-embedded tissue sections were obtained on Poly-L Lysine coated slides. Sections were then deparaffinized in xylene, hydrated in different grades of alcohol and were incubated for 1 h at room temperature with primary antibody mouse monoclonal anti SALL4 (SALL-4-EP299-rabbit monoclonal, PathnSitu, Livermore, USA) at a dilution of 1:100. Three percent hydrogen peroxide was used for endogenous peroxidase blocking. Sections were visualized by diamino benzidine (DAB) tetra hydrochloride (DAB) and counterstained with Mayer’s hematoxylin. The primary antibody was replaced by rabbit immunoglobulin G isotype control (invitrogen); blood vessels were taken as internal antigen-positive control for SALL4. Dysgerminoma was taken as a positive control for SALL4 [Figure 1].
 |
Figure 1: Strong expression of spalt.like transcription factor 4 in dysgerminoma (positive control, immunohistochemistry, 20 × 100) Click here to view |
Evaluation of slides
The slides were evaluated using a light microscope, OLYMPUS BLX4 (Tokyo, Japan). The expression of SALL4 was considered positive based on the prominent brown nuclear staining taken up by the nucleus, cytoplasm, and both of these. In each case, three fields were randomly selected and evaluated. The percentages of positive tumor cells were evaluated in a semiquantitative manner by two observers independently.
Staining intensity
SALL4 expression was evaluated semiquantitatively, and the intensity was graded as 0 – no expression, 1 – weak expression, 2 – moderate expression, and 3 – strong expression.
The extent or proportion of tumor cells
The degree of positive tumor cells for SALL4 was graded as 0: no labeling; 1: ≤30%; 2: 31%–60%; 3: >60%. Further scoring was evaluated by multiplying the score of staining intensity and extent (0–9).[19] The overall score was subjected to further survival analysis.
Statistical analysis
The Statistical Package for the Social Sciences version 21.0, IBM, USA was used for statistical analysis to study the association of the clinicopathological correlation and expression of SALL4, the comparison and association within the groups (leukoplakia with dysplasia and OSCC) using Chi-square test and between these two groups, Kruskal–Wallis test estimated the overall significance followed by Bonferroni post hoc test. Kaplan–Meier test analyzed cumulative overall survival. Log-rank test analyzed the relationship between grade and survival [Figure 2]. A value of P = 0.05 was considered statistically significant.
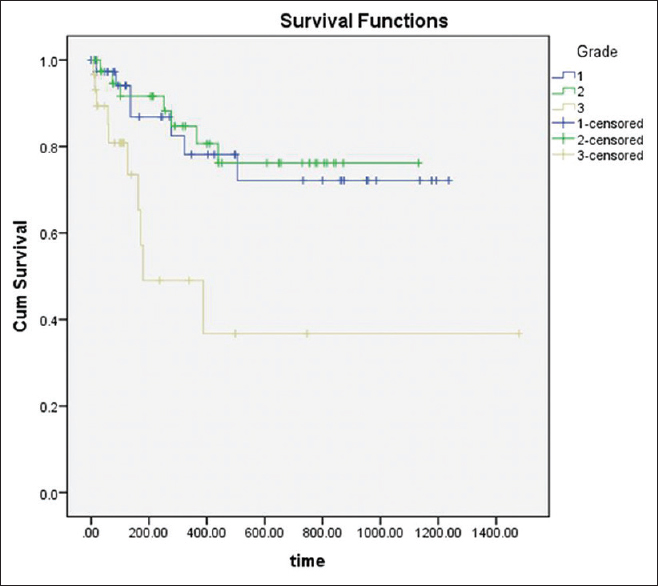 |
Figure 2: Kaplan–Meier survival curves illustrating the significance of spalt-like transcription factor 4 expression in various grades of oral squamous cell carcinoma. Poorly differentiated oral squamous cell carcinoma had significantly reduced 46.6 months survival rate Click here to view |
Results
The demographic details of patients of OSCC have been tabulated in [Table 1]. SALL4 was observed in 100/110 cases, whereas 6 cases of Stage I and 4 cases of Stage II were SALL4 negatives. Overexpression of SALL4 was observed in poorly differentiated OSCC when compared to moderately and well-differentiated carcinoma [Table 2]; P = 0.001; [Figure 3]. Although most of the cases of well-differentiated OSCC stained for SALL4, the intensity of staining was weak [Figure 3]. A significant level of overexpression of the protein (Immuno Reactive Score [IRS] of 6 and above) was observed in Stage IV when compared to Stage I and II [Table 2]; P = 0.001]. Patients with IRS of 6 and above in Stage I (5 cases) recurred, and similar findings were observed with well-differentiated cases with metastasis (4 cases) while in leukoplakia with dysplasia, the SALL4 expression was weak with a range of 2–4. Leukoplakia with dysplasia, moderate dysplasia (12/15), and severe dysplasia 11/15 (74%) cases exhibited weak expression for SALL4 with IRS between 2 and 4 [Figure 4]; P = 0.8] with no statistical significance between the two groups (moderate and severe). Seven cases of dysplasia were transformed into SCC. The time interval for transformation ranged between 6 months and 2 years.
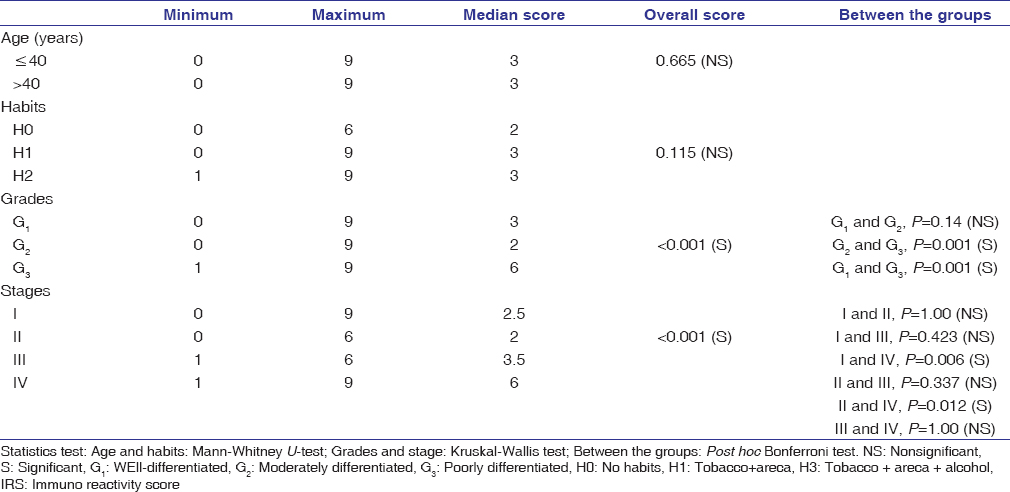 |
Table 2: Association between clinicopathological correlation and Spalt.like transcription factor 4 immuno reactive score Click here to view |
| Figure 3: Spalt-like transcription factor 4 expression: Strong in poorly differentiated squamous cell carcinoma (a); Weak to moderate expression in well-differentiated squamous cell carcinoma (b); Strong expression in well differentiated squamous cell carcinoma with metastasis (c) (immunohistochemistry, 20 × 100) Click here to view |
 |
Figure 4: Spalt-like transcription factor 4 expression: Weak in moderate dysplasia (a); Moderate in severe dysplasia (b) (immunohistochemistry, 20 × 100) Click here to view |
Survival analysis demonstrated that overexpression of SALL4 was associated with a lower survival rate (46.6 months) for poorly differentiated OSCC [log-rank test, P < 0.001; [Figure 3], indicating that SALL4 protein expression is a useful biomarker for prognostication.
Human SALL4 gene localizes on chromosome 20q13.13-q13.2, a member of the SALL gene family, and acts as a zinc-finger transcription factor. This molecular marker is downregulated in a fully differentiated cell, with the maturation of the tissues, the level of SALL4 decreases. Similar were the findings in our control group, i.e., the expression of SALL4 was observed in the few basal cells suggesting the pluripotential nature. Loss of these genes in humans is known to cause an autosomal dominant disease, Okihiro syndrome (duane radial ray syndrome), which is associated with multiple organ defects.[20] The mechanism by which SALL4 is ubiquitinated is yet to be identified, however, one of them is by the ubiquitin-proteasome system of an E3 ubiquitin-protein ligase, tripartite motif-containing 21 has been identified.[21] Others include autoregulation by SALL4 protein suppressing SALL4 transcription.[22] Various posttransitory mechanisms for SALL4 stability include MicroRNAs viz. MicroRNA-107, microRNA-33b, SUMOylation on the lysine residue, nuclear receptor-binding protein 1 regulation of SALL4 protein.[23],[24],[25],[26]
The demographic details of patients included are described in [Table 1]. Positive expression of SALL4 was seen in OSCC (100 cases) with heterogeneous expression with few cells exhibiting the absence of staining. Leukoplakia with dysplasia (thirty cases) was also positive to SALL4 with variation in the staining and intensity. Although 7 cases of dysplasia were converted to OSCC having an IRS score between 2 and 4, further studies are needed to understand the pathogenesis of the disease process. The brown color that is considered positive expression for SALL4 was seen in the cytoplasm as well in the nucleus, though its positivity is mainly observed in the nucleus. This could be related to mutated (lysine 64 was mutated into arginine) form of SALL4, which does not completely lose its trans-activating capability in the cells, thus detected in the nucleus. It can also be speculated depending on the various stimulant or signaling factors, these molecules can switch between the nucleus and cytoplasm, which requires further investigation.[27],[28],[29]
Overexpression of SALL4 in cancer and its action on multiple cellular processes that are involved in tumor initiation, proliferation, migration, and invasion has been proved in various lesions. It regulates the proliferation of cells through beta-catenin/cyclin D1, BMI, and PTEN pathways. SALL4 also binds to the HOXA9, FADD, and BMI-1, which are the promoter regions of the genes involved in apoptosis.[30],[31] As FADD has an important role in apoptosis, its loss can give cancer cells the proliferative advantage as apoptosis would no longer be induced. The self-renewal of CSCs by SALL4 is regulated through Oct4, Nanog, Sox-2 pathways.[32] The study was in concordance with the studies on breast and lung cancer.[9],[33],[34] but did vary from other researchers.[29]
It has been observed that SALL4 upregulates Wnt/β-catenin signaling pathway by directly binding to catenin beta1 promoter and trans-activating catenin beta1 leading to self-renewal and pluripotency of embryonic stem cells and inhibit stem cell differentiation.[10] Overexpression of this molecule was also observed in endometrial carcinoma.[24] This study connoted SALL4 binding to the c-Myc promoter region and activating the SALL4/c-Myc pathway, which could activate tumorigenesis. He et al. in 2013 found wild type-epidermal growth factor (EGF), which is activated by EGF ligand, roots the upregulation of SALL4 in lung tumor.[23] Li et al. in 2015 found that SALL4 was not expressed in normal and hyperplastic tissues.[10] Few cases in moderate and severe dysplasia in the present study did show weak expression. On the contrary, 7/30 cases of dysplasia were transformed into SCC in the span of 3 years, which would need further investigation. Since these patients were under close follow-up, appropriate treatment was rendered and are now disease-free.
In this study, we found SALL4 was significantly higher in poorly differentiated OSCC, which was in concordance with studies on breast carcinoma, which showed that high expression of SALL4 is associated with advanced tumor invasion compared to the normal adjacent tissue.[9] Expression of SALL4 was evaluated by Ota et al. in 2015, who found higher expression of this marker in OC compared to the normal mucosa and leukoplakia and concluded that it might play a role in cell proliferation and can serve as a therapeutic target.[12] Elevated SALL4 has shown to upregulate the expression of twist1 and N-cadherin, while the expression of E-cadherin is downregulated, thereby prompting epithelial-mesenchymal transition (EMT). By acting on E-cadherin, it releases the intercellular adhesion leading to cellular motility. Furthermore, the EMT factor ZEB1 is upregulated by SALL4.[20] Further, studies done by Ram et al. in 2017 have identified three CSCs populations in moderately differentiated lip squamous cell carcinoma (SCC), one of which being SALL4 and they also found that high expression of SALL4 could be associated with poor survival of patients.[14] Similar findings were also seen in moderately differentiated SCC of buccal mucosa in our study, thus stating CSCs may be a potential novel therapeutic target.[13]
Four cases of well-differentiated SCC with metastasis in our study had exhibited higher expression for SALL4. Five cases of Stage I and well-differentiated cases with metastasis (4 cases) also exhibited higher expression (IRS score 6 and above), and these patients presented with recurrence, thus confirming SALL4 as a prognostic marker. Recurrence was seen in 9 patients: buccal mucosa (5) and tongue (4), and metastasis were seen in the regional lymph nodes [Table 1] and [Table 2].
The limitation of this study was that we were not able to categorize high and low expression for SALL4 since the samples in each category were limited. Literature review shows that these stemness markers are resistant to chemo and radiotherapy due to over-expression of various molecular determinants, major ones being adenosine triphosphate binding cassette (ABC) transporters (ABC) as they cause drug efflux and low drug concentrations. A small population of CSCs known as side population cells along with increased ABC transporters are also known to contribute to tumor relapse.[20] The suppression of ABC transporters increases anticancer drug sensitivity in cancers.
Conclusions
Overexpression of SALL4 was strongly expressed in poorly differentiated and well-differentiated OSCC with metastasis suggesting it being a marker of prognostication and thus could serve as a potential therapeutic target in OC. However, further studies with a panel of stem cell markers, antiapoptotic, and proliferative invasive markers along with stromal environment with increased sample size will help to understand the molecular mechanism involved in the tumorigeneses especially cases of severe dysplasia which has predilection to transform into SCC and to prove it as one of the prognostic marker.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
| 1. | |
| 2. | |
| 3. | |
| 4. | |
| 5. |
Zhao W, Li Y, Zhang X. Stemness-related markers in cancer. Cancer Transl Med 2017;3:87-95.
 [Full text] |
| 6. | |
| 7. | |
| 8. | |
| 9. | |
| 10. | |
| 11. | |
| 12. | |
| 13. | |
| 14. | |
| 15. | |
| 16. | |
| 17. | |
| 18. | |
| 19. | |
| 20. | |
| 21. | |
| 22. | |
| 23. | |
| 24. | |
| 25. | |
| 26. | |
| 27. | |
| 28. | |
| 29. | |
| 30. | |
| 31. | |
| 32. | |
| 33. | |
| 34. |
