Xiangshan Zhao1, Gautam K Malhotra1, Hamid Band2, Vimla Band3
1 Department of Genetics, Cell Biology and Anatomy, University of Nebraska Medical Center, Nebraska Medical Center, Omaha, NE, USA
2 Department of Genetics, Cell Biology, Anatomy, Biochemistry, Molecular Biology, Pathology, Microbiology, Pharmacology and Experimental Neuroscience, College of Medicine, and the Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Nebraska Medical Center, Omaha, NE, USA
3 Department of Genetics, Cell Biology and Anatomy; Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Nebraska Medical Center, Omaha, NE, USA
| Date of Submission | 16-Sep-2011 |
| Date of Acceptance | 20-Oct-2011 |
| Date of Web Publication | 31-Dec-2011 |
Correspondence Address:
Vimla Band
Department of Genetics, Cell Biology and Anatomy; Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Nebraska Medical Center, Omaha, NE
USA
Source of Support: None, Conflict of Interest: None
DOI: 10.4103/1477-3163.91415
Abstract
Introduction: Emerging evidence suggests a direct role of cancer stem cells (CSCs) in the development of breast cancer. In vitro cellular models that recapitulate properties of CSCs are therefore highly desirable. We have previously shown that normal human mammary epithelial cells (hMECs) immortalized with human telomerase reverse transcriptase (hTERT) possess properties of mammary stem / progenitor cells. Materials and Methods: In the present study, we used this cell system to test the idea that other known hMEC-immortalizing oncogenes (RhoA, HPVE6, HPVE7, p53 mutant, and treatment with g-radiation), share with hTERT, the ability to maintain mammary stem / progenitor cells. Results: The results presented here demonstrate that similar to hMECs immortalized with hTERT, all hMEC cell lines immortalized using various oncogenic strategies express stem / progenitor cell markers. Furthermore, analyses using 2D and 3D culture assays demonstrate that all the immortal cell lines retain their ability to self-renew and to differentiate along the luminal lineage. Remarkably, the stem / progenitor cell lines generated using various oncogenic strategies exhibit a block in differentiation along the myoepithelial lineage, a trait that is retained on hTERT-immortalized stem / progenitors. The inability to differentiate along the myoepithelial lineage could be induced by ectopic mutant p53 expression in hTERT-immortalized hMEC. Conclusions: Our studies demonstrate that stem / progenitor cell characteristics of hMECs are maintained upon immortalization by using various cancer-relevant oncogenic strategies. Oncogene-immortalized hMECs show a block in their ability to differentiate along the myoepithelial lineage. Abrogation of the myoepithelial differentiation potential by a number of distinct oncogenic insults suggests a potential explanation for the predominance of luminal and rarity of myoepithelial breast cancers.
Keywords: Immortalization, luminal differentiation, myoepithelial differentiation, self-renewal, stem cell
How to cite this article:
Zhao X, Malhotra GK, Band H, Band V. A block in lineage differentiation of immortal human mammary stem / progenitor cells by ectopically-expressed oncogenes. J Carcinog 2011;10:39
How to cite this URL:
Zhao X, Malhotra GK, Band H, Band V. A block in lineage differentiation of immortal human mammary stem / progenitor cells by ectopically-expressed oncogenes. J Carcinog [serial online] 2011 [cited 2021 Oct 14];10:39. Available from: https://carcinogenesis.com/text.asp?2011/10/1/39/91415
Introduction
Breast cancer is the most common malignancy and the second leading cause of cancer-related deaths among women in the United States. [1] Breast cancer is now recognized as a genetically and clinically heterogeneous disease. [2] However, it remains unclear whether the oncogenic transformation of distinct target cells within the mammary gland contribute to this heterogeneity, nor is it clear as to which cell types are most susceptible to oncogenesis.
According to the classical theories of carcinogenesis, mammary tumor formation is thought to be a consequence of cumulative genetic alterations in any epithelial cell within the mammary gland and all cells are considered equally likely to serve as cells of origin for breast cancers. In recent times, however, the detection of cells that share phenotypic markers as well as genetic programs of normal stem / progenitor cells has led to the emergence of the cancer stem cell (CSC) hypothesis, challenging the long-held and well-accepted prior paradigm. According to the CSC hypothesis, genetic alterations in normal stem / progenitor cells deregulate mechanisms that ensure normal stem / progenitor homeostasis and such transformed cells acquire the key attributes of cells of origin of cancer, such as, continuous proliferation, invasion, and metastasis. [3],[4],[5],[6],[7] Alternatively, CSCs can arise by acquisition of stem-like properties upon oncogenic transformation of more differentiated cells. Similar to normal stem cells, the CSCs retain the ability to self-renew and a certain degree of cellular differentiation; expansion of CSCs is thought to facilitate tumor progression, whereas, differentiation is thought to contribute to phenotypic heterogeneity in tumors.
Although the cancer stem cell hypothesis remains controversial, there is a growing body of evidence to support it. However, the molecular mechanisms by which normal stem cells become transformed into CSCs remain largely unknown. Therefore, it is critical to establish and characterize cellular models that can allow analyses of molecular mechanisms that control self-renewal and differentiation of both normal and cancer stem cells.
Of late, expression profiling of tumors has identified six major molecular subtypes of breast cancer: basal epithelial-like, ErbB2-overexpressing, normal breast epithelial-like, luminal epithelial subtypes A and B, and claudin low; patients corresponding to these different subtypes carry significantly different survival and treatment outcomes. [8],[9],[10] The correspondence of some breast cancer subtypes with cell types present in the normal mammary gland (such as basal and luminal) strongly supports the idea that breast tumor subtypes may represent malignancies of biologically distinct cell subtypes. The relatively rare occurrence of tumors with myoepithelial cell characteristics (or myoepithelioma), tumors with a much worse prognosis, [11] suggests that oncogenesis of precursor cells that results in common subtypes of breast cancer may be associated with abrogation of the program of differentiation toward myoepithelial lineage.
We have previously shown that normal mammary epithelial cells (hMECs) isolated from human reduction mammoplasty specimens using the DFCI-1 medium exhibit stem / progenitor properties; importantly, these cells retain their stem / progenitor cell characteristics of self-renewal and differentiation toward luminal and myoepithelial cell lineages, even when immortalized with hTERT. [12] In other studies, we have demonstrated that a number of human cancer-associated, known or putative oncogenes, such as p53 mutant, [13] RhoA, [14] and papilloma virus protein E6 (HPVE6) and / or E7 (HPVE7), [15],[16] as well as treatment with g-radiation, can immortalize hMECs. [17] Utilizing these distinct cellular models, we have investigated the impact of various immortalizing oncogenes on hMEC stem / progenitor cell self-renewal and differentiation toward luminal and myoepithelial lineages. These analyses reveal quite striking results, while cells immortalized using all of the tested oncogenic strategies maintain self-renewal as well as varying degrees of differentiation toward the luminal lineage, their ability to differentiate toward myoepithelial lineage is completely abrogated. In contrast, TERT-immortalized hMECs retain the ability to differentiate into both the myoepithelial and luminal lineages. These results suggest that disruption of differentiation toward myoepithelial lineage is an early event upon oncogenic transformation of the human mammary stem / progenitor cells.
Materials and Methods
Isolation of normal mammary epithelial cells
The normal mammary epithelial cell strain 76N was isolated from a reduction mammoplasty specimen and grown in DFCI-1 medium, as described previously. [16]
Immortalization of normal mammary epithelial cells
76N cells were immortalized by ectopic overexpression of hTERT, human p53 mutant (del239), [13] human papillomavirus oncoprotein E6 and / or E7, [15] or human RhoA, [14] or by treatment with γ-radiation, [17] and the immortal cells were cultured in DFCI-1 medium, as described previously. [13],[14],[15],[16],[17],[18] The nomenclature of the 76N-derived immortal cell lines was as follows: 76N.TERT (hTERT-immortalized); 76N.p53delta239 (p53 mutant del239-immortalized); 76N.E6, 76N.E7, and 76E6E7 (HPV16 E6- , E7- or E6 / E7-immortalized); 76N.RhoA.WT (wild type human RhoA-immortalized); and 76N.R30 (g-radiation-immortalized). Immortalized hMEC lines were maintained in DFCI-1 medium with appropriate drug selection as described. [16]
Overexpression of mutant p53 in 76N.TERT cells
A lentiviral expression construct pLenti6-p53-R249S coding for the human p53 mutant R249S was obtained from Addgene. Empty vector or mutant p53 constructs were transfected into 293FT packaging cells to generate lentivirus supernatants for infection of 76N.TERT cells, and stable transductants were selected in 15 μg / ml blasticidin.
Western blot analysis
Cell lysate preparation and western blotting, using the indicated antibodies, were performed as previously described. [12] The cell lysate protein concentrations were quantified using the BCA protein assay kit (Pierce).
Self-renewal and differentiation analyses
The indicated immortal cells were grown in serum-free MEGM medium (Lonza Group Ltd.) supplemented with B27 (10 ml / 500 ml medium, Invitrogen), 20 ng / ml each of EGF (Invitrogen) and bFGF (BD Biosciences), and 4 μg / ml heparin (Sigma), without bovine pituitary extract. [12],[19] Once the 76N.TERT-cells exhibited evidence of differentiation (about 30 days), the immortal cell lines under examination were re-plated on coverslips and analyzed for expression of various markers using immunofluorescence staining.
Matrigel culture
Glass coverslips placed in 24-well plates were coated with Matrigel (BD Bioscience), normal and immortal cells (2000 cells / well were plated on Matrigel-coated coverslips in DFCI-2 (D2) medium [15],[16] containing 2% Matrigel). [20],[21] After 12 days of culture, the cells were fixed and used for immunofluorescence staining. [21]
Immunofluorescence staining
Cells cultured on 2D (for MEGM culture) or 3D (Matrigel-coated with D2 media) coverslips were fixed in 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 and blocked in 5% donkey serum. Immunostaining was carried out by two to three hours of incubation, with optimal concentrations of the indicated antibodies, followed by Alexa Flour 594-conjugated donkey anti-mouse (1 : 1,000) or donkey anti-rabbit (1 : 1,000) antibodies or Alexa Flour 488-conjugated donkey anti-rabbit (1:1,000) antibodies for one hour. Mouse anti-human α-smooth muscle actin (α-SMA)-Cy3 was used for α-SMA staining. [12] The slides were mounted using ProLong; Gold Antifade (Invitrogen) and images were acquired under a Zeiss Axioplan fluorescence microscope, using a 20X objective (for MEGM cultured cells) or an LSM510 fluorescence confocal microscope (Carl Zeiss, Germany), using a 63X objective (for Matrigel cultured cells).
Results
Normal hMECs isolated and cultured in DFCI-1 medium and their immortalized derivatives express mammary stem / progenitor cell markers
We have previously shown that a normal hMEC strain 70N and its hTERT-immortalized derivative (70N.TERT) exhibit stem / progenitor cell properties. [12] We have also previously shown that an independently-derived normal hMECs strain (76N), obtained from a separate reduction mammoplasty specimen, [15],[16] is highly susceptible to immortalization using a number of oncogenes / cellular genes, overexpression of hTERT, or treatment with γ-irradiation. [13],[14],[15],[16],[17],[18] Western blot analyses showed that 76N as well as its immortal derivatives express mammary stem cell (K5, K14, ALDH1A3, p63, CD29) as well as luminal cell (K18, E-cadherin) markers [Figure 1]a. Immunofluorescence staining showed that these cells also express the stem cell marker CD49f [Figure 1]b. These results suggest that, similar to TERT-immortalized hMECs, immortal cells obtained using various oncogenes / cellular genes as well as those immortalized with γ-irradiation may exhibit stem / progenitor cell properties.
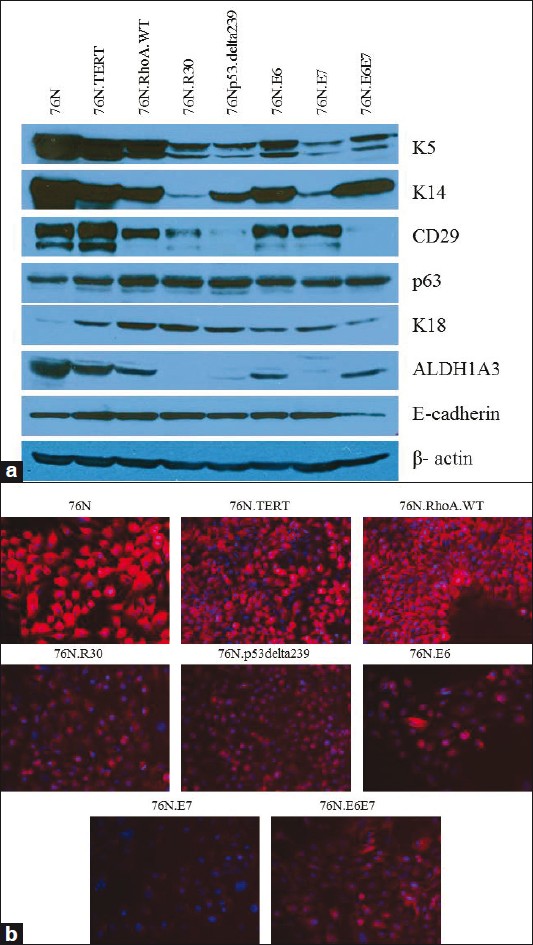 |
Figure 1: Analyses of stem / progenitor cell markers in parental and immortalized human mammary epithelial cells isolated and cultured in the DFCI-1 medium, (a) Western blotting of parental and immortal cells. A total of 50 μg of cell lysates were western blotted, using indicated antibodies. β -actin was used as loading control, ( b ) Immunofluorescence staining of normal and immortal hMECs using CD49f antibody (red), and DAPI (blue) used for nuclear staining is shown ×20 Click here to view |
All immortalized derivatives of 76N exhibit self-renewal and an ability to differentiate into luminal lineage cells
We have previously shown that immortalized human mammary stem / progenitor cells grown in DFCI-2 (D2) medium form compact colonies in which a proportion of cells stained for stem / progenitor markers, reflecting self-renewal, while other cells showed evidence of luminal cell differentiation. [12] Notably, 76N-TERT and other immortal derivatives of 76N cell strain also exhibited a compact colony growth in the D2 medium (data not shown). To directly assess the self-renewal and luminal differentiation of immortalized 76N derivatives, we first used the differentiation induction protocol in a two-dimensional (2D) culture, with cells grown in the MEGM medium, followed by staining for the stem / progenitor cell marker K5 (stained in green) and the luminal cell marker MUC1 (stained in red) [Figure 2]a. All the immortal cell lines grown under these conditions showed a mixed phenotype, with distinct K5-staining and MUC1-staining cells [Figure 2]a; only rarely these cells were positive for both markers, while a substantial proportion of cells (that were positive for DAPI) were negative for both markers, likely reflecting intermediate progenitors or partially differentiated cells. Thus, similar to our previous findings with the TERT-immortalized 70N cell line, mammary stem / progenitor cell lines, immortalized using various oncogenic strategies, retained their ability to self-renew and to differentiate along the luminal lineage.
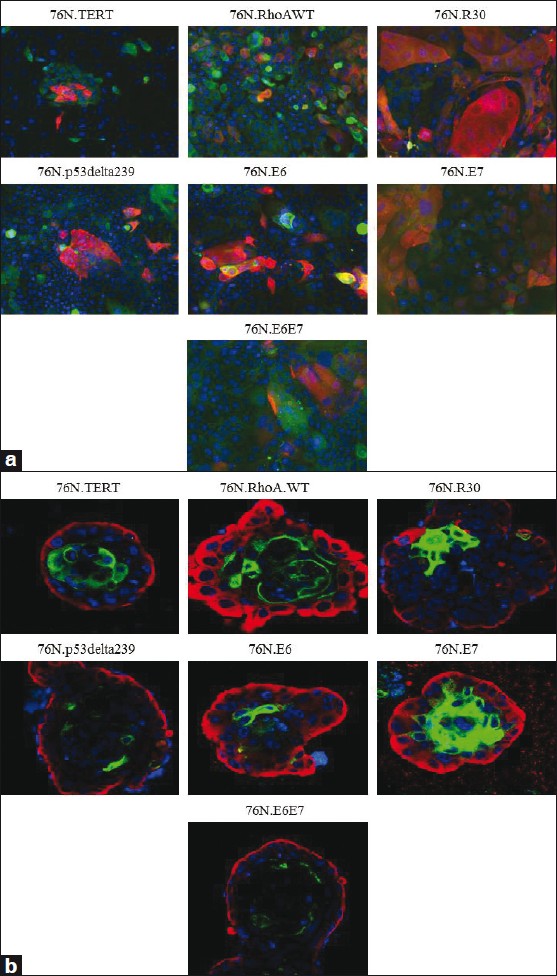 |
Figure 2: In vitro self-renewal and luminal differentiation of immortalized hMECs. (a) Cells cultured in a two-dimensional culture with MEGM media. Immunostaining of cells with rabbit-anti human K5 (green) and mouse anti-human MUC1 (red) ×20, (b) Cells cultured in three-dimensional Matrigel with DFCI-2 medium. The acini were co-stained with rabbit-anti human K5 (red) and FITC conjugated anti-human MUC1 antibody (green) ×63 Click here to view |
Others have shown that mammary stem / progenitor cells in three-dimensional (3D) Matrigel culture showed evidence of differentiation. [22] We therefore cultured different immortal cells in 3D Matrigel, in the D2 medium for 12 days and analyzed them, by immunostaining for K5+ stem / progenitor cells (red) and MUC1+ cells (green), which represented luminal-differentiation [Figure 2]b. Indeed, each cell line showed an outer layer of K5+ / MUC1- cells, consistent with the self-renewal of stem / progenitor cells during acinus formation; in contrast, cells near the lumens of the acini were K5- / MUC1+, indicating their differentiation along the luminal lineage [Figure 2]b. Together, our results using two different protocols clearly demonstrated the ability of hMEC stem / progenitor cells generated using different immortalizing strategies to self-renew as well as to maintain their ability to differentiate into the luminal lineage, albeit to varying degrees.
Mammary stem / progenitor cells immortalized using oncogenic strategies other than hTERT show a block in differentiation into the myoepithelial lineage
As we established earlier, hTERT-immortalized mammary stem progenitor cells are capable of in vitro differentiation along the myoepithelial lineage. [12] To examine if immortal stem / progenitor cell derivatives of 76N cells obtained using other oncogenes were capable of myoepithelial lineage differentiation, we cultured these cells in MEGM medium. A morphological characteristic of cells that differentiated along the myoepithelial lineage in this system was the characteristic localization of relatively elongated myoepithelial cells organized in a loose pattern around the perimeter of the compact colonies harboring self-renewing cells, and those cells that underwent luminal differentiation. Indeed, the 76N.TERT cell line showed this characteristic pattern [Figure 3]a. In contrast, the stem / progenitor cell lines established using other oncogenic modalities failed to exhibit this pattern; instead, these cell lines exhibited colonies without surrounding myoepithelial cells [Figure 3]a. Complementing the morphological evidence, the peripheral elongated cells seen in the TERT-immortalized cell cultures showed many cells that were positive for myoepithelial marker α-smooth muscle actin (α-SMA), while centrally located cells were expectedly negative for this marker [Figure 3]b. In contrast, no α-SMA+ cells were observed in cultures of other immortalized stem / progenitor cell lines [Figure 3]b.
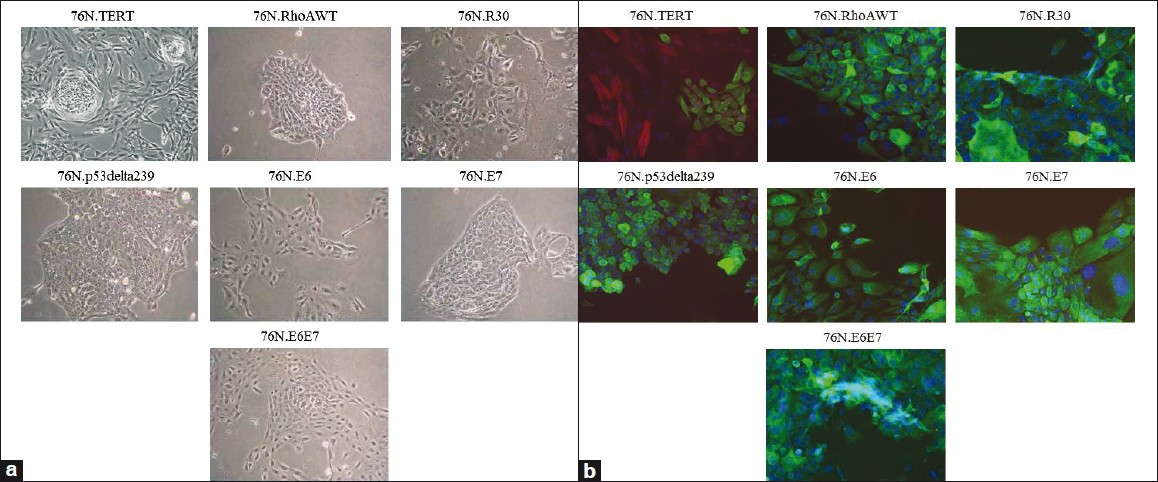 |
Figure 3: In vitro self-renewal and myoepithelial cell differentiation of immortalized hMECs in MEGM medium, (a) Morphology of cells after beginning of differentiation, ×10 (b) Immunofluorescence staining of myoepithelial cells. The cells were co-stained with rabbit-anti human K5 (green) and a myoepithelial cell marker (α -SMA), with mouse anti-human α -SMA antibody (red) ×20 Click here to view |
Introduction of a mutant p53 into the TERT-immortalized mammary stem / progenitor cell line blocks its ability to differentiate toward the myoepithelial lineage
The inability of the stem / progenitor lines, analyzed earlier, to differentiate along the myoepithelial lineage could represent a selection of precursors that no longer retain this differentiation potential or an active suppression of the potential for myoepithelial cell differentiation by the oncogenes used. To begin to address these possibilities, we assessed the impact of the ectopic expression of a p53 mutant, R249S, on the myoepithelial differentiation-competent 76N.TERT stem / progenitor cell line. The overexpression of the introduced p53 was confirmed by western blotting [Figure 4]a. Analyses using the morphological assay of differentiation upon a culture in an MEGM medium indicated that the vector-infected cells retained their ability to form differentiated myoepithelial cells surrounding the central colonies [Figure 4]b; in contrast, the 76N.TERT cells expressing the p53-R249S mutant showed a lack of peripheral cells with myoepithelial morphology, and instead showed only compact colonies [Figure 3]b. These results support the likelihood that suppression of myoepithelial cell differentiation is dominantly induced by oncogenes such as mutant p53, and this behavior is unlikely to be due to a chance selection of precursors that lack such differentiation potential.
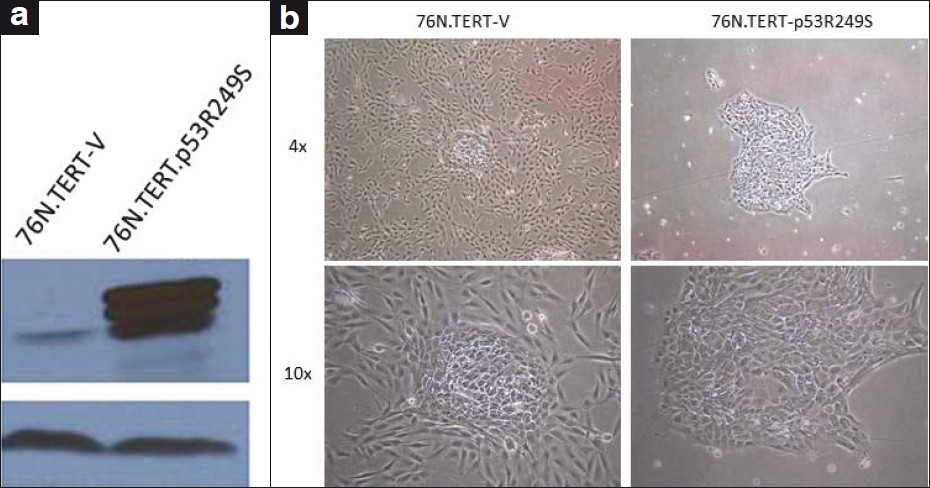 |
Figure 4: Effect of p53 mutant on 76N.TERT myoepithelial cell differentiation (a) Western blotting shows mutant p53 overexpression, (b) Lack of myoepithelial cell differentiation in 76N.TERT-p53R249S compared with 76N.TERT-V. Images were acquired under a Nikon inverted microscope using ×4 and ×10 objectives Click here to view |
Discussion
Cancer stem cell hypothesis is increasingly accepted as a paradigm for tumor initiation and maintenance. Yet, it remains unclear why tumors that presumably initiate in stem / progenitor cells do not produce tumors or different lineages relatively uniformly. This discrepancy is particularly obvious in breast cancer, where a majority of tumors represent a luminal lineage (ER+ as well as ErbB2+ tumors), while tumors representing the myoepithelial lineage are distinctly rare. One potential explanation for this paradox is that differentiation into the myoepithelial lineage is blocked during oncogenic transformation of stem / progenitor cells. Here, we have used a unique immortal human mammary stem / progenitor cell system, which we have recently shown to exhibit the critical stem cell traits of self-renewal as well as luminal and myoepithelial differentiation in vitro, to address this issue. Our analyses show that human mammary stem / progenitor cells rendered immortal using hTERT (as in our previous studies) retain both self-renewal and differentiation along luminal and myoepithelial lineages. In contrast, a number of distinct oncogenic insults lead to immortalized derivatives that retain the self-renewal potential and differentiation along the luminal lineage, but all oncogenic modalities uniformly block differentiation along the myoepithelial lineage.
The oncogenic modalities that we tested included HPV E6 / E7, which are known to be pathophysiological relevant to other epithelial cell systems, but are used as model oncogenes in mammary oncogenesis, as their target pathways (such as Rb and p53) are directly implicated in mammary oncogenesis. In addition, we have used strategies that directly model breast cancer-associated oncogenic insults, including the expression of mutant p53 genes, fractionated γ-irradiation, and overexpression of the small GTPase RhoA.
Regardless of the genetic insult used to promote immortalization, the myoepithelial differentiation potential of hMECs was abrogated. Importantly, when a mutant p53 was overexpressed in cells immortalized with hTERT, the ability of the immortal parent to differentiate along the myoepithelial pathway was abrogated. Thus, it was unlikely that the various oncogenes used might selectively immortalize a naturally occurring population of progenitors that was devoid of myoepithelial differentiation. Instead, our results suggested that different oncogenic insults actively inhibit myoepithelial differentiation potential.
If the finding that oncogenic insults inhibit the ability of mammary stem / progenitor cells to differentiate along the myoepithelial lineage is validated in vivo, it could provide an explanation for why mammary tumors representing this lineage are relatively rare. If indeed this mechanism is operational in human cancer, it can be anticipated that cancer stem cells from human breast cancers may fail to differentiate along the myoepithelial lineage, while retaining the luminal differentiation potential, a possibility that will be of considerable interest, to test in future experiments.
It is not clear why oncogenic transformation of mammary stem / progenitor cells is associated with a block in the myoepithelial differentiation potential. It has long been argued that myoepithelial cells may alter oncogenesis by serving as tumor suppressors or tumor promoters. Indeed, several studies have suggested the tumor-suppressive role of myoepithelial cells. [23],[24],[25] Thus, one possibility is that a block in the myoepithelial lineage differentiation potential represents loss of an inherent tumor suppressor mechanism. In this regard, it is rather remarkable that most of the immortal hMEC cells we tested here, represent models of relatively early neoplastic transformation, as these cells (with the exception of the 76N.R30 line immortalized with γ-irradiation) do not show advanced oncogenic traits such as anchorage-independent growth or the ability to grow as xenotransplants. [17] Thus, the potential tumor suppressor mechanism exerted by myoepithelial cells may be exerted at an early stage. Our ability to isolate myoepithelial lineage cells from the same parental stem / progenitors (immortalized using hTERT) must allow future co-culture studies and co-implantation in xenotransplant experiments, to directly test if this line of reasoning based on the literature can explain our experimental findings. A potential explanation for our findings is that the oncogene-immortalized mammary stem / progenitor cells, in contrast to the hTERT-immortalized ones, lack the ability to utilize certain growth factors needed for the in vitro growth of myoepithelial progeny or that they use factors that inhibit myoepithelial differentiation. For example, very recently it has been shown that the epidermal growth factor (EGF) causes a massive expansion of the myoepithelial lineage, whereas, amphiregulin is required for normal ductal development. [26] As the in vitro culture requirements for luminal versus myoepithelial cell lineages, especially of human mammary cell origin, remain undefined, further studies are necessary to assess the possibility that loss of myoepithelial lineage may represent loss of particular growth factors that have not been identified. However, it should be testable as culture systems are further adapted.
The ability to immortalize human mammary stem / progenitors with full or partial differentiation potential provides a unique ability not only to assess the importance of altered differentiation programs (as discussed earlier) in mammary oncogenesis, but also to begin to examine the molecular pathways associated with lineage differentiation in a human mammary cell system. In this regard, we have now established conditions that allow the whole genome mRNA and microRNA expression analyses to be performed in this system. Thus, future studies, using the cellular models described here, should help provide answers to the key questions related to mammary stem cell biology and oncogenesis.
Acknowledgment
The study in our laboratories is supported by the NIH grants CA96844 and CA144027 to VB, CA87986, CA99163, CA105489, CA116552, and NCI 5U01CA151806-02 to HB, Department of Defense grants W81XWH-07-1-0351 and W81XWH-11-1-0171 to VB; and the NCI Core Support Grant to the UNMC-Eppley Cancer Center. Gautam Malhotra is supported by the UNMC Graduate Student Fellowship.
References
| 1. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin 2010;60:277-300.  [PUBMED] [FULLTEXT] |
| 2. | Morrow PK, Hortobagyi GN. Management of breast cancer in the genome era. Annu Rev Med 2009;60:153-65.  [PUBMED] [FULLTEXT] |
| 3. | Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol 2009;27:44-6.  [PUBMED] [FULLTEXT] |
| 4. | Charafe-Jauffret E, Monville F, Ginestier C, Dontu G, Birnbaum D, Wicha MS. Cancer stem cells in breast: Current opinion and future challenges. Pathobiology 2008;75:75-84.  [PUBMED] [FULLTEXT] |
| 5. | Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res 2009;69:1302-13.  [PUBMED] [FULLTEXT] |
| 6. | Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol 2007;23:675-99.  [PUBMED] [FULLTEXT] |
| 7. | Dalerba P, Cho RW, Clarke MF. Cancer stem cells: Models and concepts. Annu Rev Med 2007;58:267-84.  [PUBMED] [FULLTEXT] |
| 8. | Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010;12:R68.  [PUBMED] [FULLTEXT] |
| 9. | Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001;98:10869-74.  [PUBMED] [FULLTEXT] |
| 10. | Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406:747-52.  |
| 11. | Hungermann D, Buerger H, Oehlschlegel C, Herbst H, Boecker W. Adenomyoepithelial tumours and myoepithelial carcinomas of the breast–A spectrum of monophasic and biphasic tumours dominated by immature myoepithelial cells. BMC Cancer 2005;5:92.  [PUBMED] [FULLTEXT] |
| 12. | Zhao X, Malhotra GK, Lele SM, Lele MS, West WW, Eudy JD, et al. Telomerase-immortalized human mammary stem / progenitor cells with ability to self-renew and differentiate. Proc Natl Acad Sci U S A 2010;107:14146-51.  [PUBMED] [FULLTEXT] |
| 13. | Gao Q, Hauser SH, Liu XL, Wazer DE, Madoc-Jones H, Band V. Mutant p53-induced immortalization of primary human mammary epithelial cells. Cancer Res 1996;56:3129-33.  [PUBMED] [FULLTEXT] |
| 14. | Zhao X, Lu L, Pokhriyal N, Ma H, Duan L, Lin S, et al. Overexpression of RhoA induces preneoplastic transformation of primary mammary epithelial cells. Cancer Res 2009;69:483-91.  [PUBMED] [FULLTEXT] |
| 15. | Wazer DE, Liu XL, Chu Q, Gao Q, Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci U S A 1995;92:3687-91.  [PUBMED] [FULLTEXT] |
| 16. | Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc Natl Acad Sci U S A 1990;87:463-7.  [PUBMED] [FULLTEXT] |
| 17. | Wazer DE, Chu Q, Liu XL, Gao Q, Safaii H, Band V. Loss of p53 protein during radiation transformation of primary human mammary epithelial cells. Mol Cell Biol 1994;14:2468-78.  [PUBMED] [FULLTEXT] |
| 18. | Band V. In vitro models of early neoplastic transformation of human mammary epithelial cells. Methods Mol Biol 2003;223:237-48.  [PUBMED] |
| 19. | Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem / progenitor cells. Genes Dev 2003;17:1253-70.  [PUBMED] |
| 20. | Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003;30:256-68.  [PUBMED] [FULLTEXT] |
| 21. | Dimri M, Naramura M, Duan L, Chen J, Ortega-Cava C, Chen G, et al. Modeling breast cancer-associated c-Src and EGFR overexpression in human MECs: C-Src and EGFR cooperatively promote aberrant three-dimensional acinar structure and invasive behavior. Cancer Res 2007;67:4164-72.  [PUBMED] [FULLTEXT] |
| 22. | Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW. Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev 2002;16:693-706.  |
| 23. | Sternlicht MD, Kedeshian P, Shao ZM, Safarians S, Barsky SH. The human myoepithelial cell is a natural tumor suppressor. Clin Cancer Res 1997;3:1949-58.  [PUBMED] [FULLTEXT] |
| 24. | Adriance MC, Inman JL, Petersen OW, Bissell MJ. Myoepithelial cells: Good fences make good neighbors. Breast Cancer Res 2005;7:190-7.  [PUBMED] [FULLTEXT] |
| 25. | Hu M, Yao J, Carroll DK, Weremowicz S, Chen H, Carrasco D, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell 2008;13:394-406.  [PUBMED] [FULLTEXT] |
| 26. | Pasic L, Eisinger-Mathason TS, Velayudhan BT, Moskaluk CA, Brenin DR, Macara IG, et al. Sustained activation of the HER1-ERK1 / 2-RSK signaling pathway controls myoepithelial cell fate in human mammary tissue. Genes Dev 2011;25:1641-53.  [PUBMED] [FULLTEXT] |
Authors
Dr. Xiangshan Zhao, Department of Genetics, Cell Biology and Anatomy1; Biochemistry and Molecular Biology, University of Nebraska Medical Center, 985805 Nebraska Medical Center, Omaha, NE
Mr. Gautam Malhotra, Department of Genetics, Cell Biology and Anatomy1; Biochemistry and Molecular Biology, University of Nebraska Medical Center, 985805 Nebraska Medical Center, Omaha, NE
Prof. Hamid Band, Departments of Genetics, Cell Biology and Anatomy; Biochemistry and Molecular Biology; Pathology and Microbiology; and Pharmacology and Experimental Neuroscience, College of Medicine, and the Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, 985805 Nebraska Medical Center, Omaha, NE
Prof. Vimla Band, Department of Genetics, Cell Biology and Anatomy and the Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, 985805 Nebraska Medical Center, Omaha, NE
Figures
