Sangeetha Subramaniam1, Ram Krishna Thakur2, Vinod Kumar Yadav2, Ranjan Nanda1, Shantanu Chowdhury2, Anurag Agrawal2
1 International Center for Genetic Engineering and Biotechnology, Delhi, India
2 CSIR Institute of Genomics and Integrative Biology, Delhi, India
| Date of Submission | 10-Jul-2012 |
| Date of Acceptance | 10-Aug-2013 |
| Date of Web Publication | 28-Feb-2013 |
Correspondence Address:
Anurag Agrawal
CSIR Institute of Genomics and Integrative Biology, Delhi
India
Source of Support: Gates Foundation (Ranjan Nanda) and Council of Scientific and Industrial Research (Anurag Agrawal and Shantanu Chowdhury)., Conflict of Interest: None
Abstract
Lung cancer is one of the deadliest cancers worldwide, with the highest incidence and mortality amongst all cancers. While the prognosis of lung cancer is generally grim, with 5-year survival rates of only 15%, there is hope, and evidence, that early detection of lung cancer can reduce mortality. Today, only computed tomography screening has shown to lead to early detection and reduction in mortality, but is limited by being anatomic in nature, unable to differentiate between inflammatory and neoplastic pathways, and therefore, susceptible to false positives. There is increasing interest in biomarkers for lung cancer, especially those that predict metastatic risk. Some biomarkers like DNA mutations and epigenetic changes potentially require tissue from the at-risk site; some like serum proteins and miRNAs are minimally invasive, but may not be specific to the lung. In comparison, emerging biomarkers from exhaled breath, like volatile organic compounds (VOC), and exhaled breath condensate, e.g., small molecules and nucleic acids, have the potential to combine the best of both. This mini review is intended to provide an overview of the field, briefly discussing the potential of what is known and highlighting the exciting recent developments, particularly with miRNAs and VOCs.
Keywords: Exhaled breath, lung cancer, miRNA, volatile organic compounds
How to cite this article:
Subramaniam S, Thakur RK, Yadav VK, Nanda R, Chowdhury S, Agrawal A. Lung cancer biomarkers: State of the art. J Carcinog 2013;12:3
How to cite this URL:
Subramaniam S, Thakur RK, Yadav VK, Nanda R, Chowdhury S, Agrawal A. Lung cancer biomarkers: State of the art. J Carcinog [serial online] 2013 [cited 2021 Oct 21];12:3. Available from: https://carcinogenesis.com/text.asp?2013/12/1/3/107958
Minireview
Lung cancer is one of the deadliest cancers worldwide, with the highest incidence and mortality amongst all cancers [1] While the prognosis of lung cancer is generally grim, with 5-year survival rates of only 15%, there is hope, and evidence, that early detection of lung cancer can reduce mortality. Reduced mortality has recently been shown as a consequence of low dose computed tomography (LD-CT) screening, but the effect size was modest (about 20% reduction in relative risk) and false positive rates were 96%. [2] The low efficacy is attributable in part to the fact that radiological visibility of tumors requires substantial cell mass and fast growing tumors may grow substantially between consecutive CT screening tests, which were kept 1 year apart to minimize radiation exposure and costs. It is intuitively obvious that small collections of tumor cells cannot be differentiated from similar masses of inflammatory cells using anatomic imaging approaches and probably not even by metabolic imaging methods like Fluoro-deoxy-glucose positron emission tomography (FDG-PET). Based on the presumption that early detection will lead to higher likelihood of cure, alternative cellular and molecular detection strategies such as sputum cytology and molecular biomarkers in various biological samples, have been tested for potential value in the early detection of lung cancer or prognostic and therapeutic guidance.
A number of approaches have resulted in a diverse set of molecular biomarkers for lung carcinoma, particularly in recent years with the advent of next generation sequencing technologies combined with advances in bronchoscopic and imaging techniques. These have classically been genetic, such as mutations in p53, K-ras, Rb and myc genes; [3],[4],[5],[6] epigenetic (abnormal methylation of APC, TMS1, RASSF1, p16INK4a, DAPK; or chromosomal changes (deletion in chromosome 3p which harbors several tumor suppressor genes. [7],[8],[9] Such DNA based changes are robust and not susceptible to degradation or handling related changes, unlike RNA expression. Unfortunately, the applicability of DNA markers is often limited by the need for sufficient tumor tissue, as well as limited sensitivity and specificity. Further, where DNA markers are used to guide therapy, such as epidermal growth factor receptor (EGFR) mutations, cancer cells may further mutate during treatment and require repeated tumor sampling. Recent work, where circulating tumor cells (CTC) were trapped, has provided proof-of-concept for non-invasive monitoring of tumor DNA and may be more widely used in coming years. [10],[11] However, this would not be sufficient to diagnose early limited tumors, which are unlikely to be associated with CTC. Recent evidences also show the possible role of small non-coding RNAs such mi(cro) RNA as biomarkers or targets, e.g., miR17 and miR21 clusters or let-7 as a suppressor of RAS in lung cancer. [12],[13] Unlike mRNA, miRNAs are exceptionally stable and their potential as biomarkers will be discussed further in subsequent sections.
Protein biomarkers for detection in lung cancer are typically those measurable in sera (ones that are currently clinically useful include, tissue polypeptideantigen (TSA), CYFRA-21-1 and carcinoembryonic antigen for non-small cell lung carcinoma (NSCLC), and neuron-specific enolase and progastrin-releasing peptide (ProGRP) for neuroendocrine lung carcinoma). [14],[15] Other potential ones include, plasma kalikrein B1 (KLKB1), serum amyloid A, haptoglobin-alpha-2. Tumor protein expression is important for prognostic usage and therapeutic guidance. For example, expression of (EGFR) is important in guiding therapeutic use of several drugs that are specific for the EGF receptor. [16],[17] Many of these and several other molecules are at various levels of testing in clinical/pre-clinical settings for indications ranging from diagnosis to guidance of therapy. [15] Some biomarkers can even directly lead to therapeutic advances, as seen for the oncogenic EML4-ALK fusion gene in NSCLC. [18],[19],[20],[21]
While detection of tumors is an important area for lung cancer biomarker discovery, it is equally important to determine, which tumors are likely to have metastasized, and which are likely to be limited. Detection of CTC is one such approach, but remains challenging and not clinically validated. Given that it is not the primary tumor that kills but its secondary spread, it is surprising that metastasis suppressor genes (MSG) have not received much attention as biomarkers. [22] An emerging body of work aims to fill this lacuna. To establish metastases successfully tumor cells must successfully negotiate several steps, including detachment from the primary tumor, intravasation, survival and extravasation from blood vessels into the secondary site followed by colonization of the secondary site. [22] Therefore, discovery of the first gene (NM23) that could ‘single-handedly’ mitigate the metastatic process was enigmatic. [23] Several other MSGs have been identified since, including, Breast cancer Metastasis Suppressor 1 (BRMS1), KiSS, KAI1, and Rho GDP dissociation inhibitor-2 (RhoGDI2) in different cancer types. [24],[25],[26]
In order to test association of MSG levels with lung tumor progression, we conducted large-scale meta-analysis of 23 MSGs in lung tumor clinical samples available from expression project for oncology (expO) database (GSE2109; expO is hosted by International Genomics Consortium (IGC, USA, www.intgen.org )). Interestingly, investigation of 93 patient derived lung tumor transcriptomes grouped stage wise showed statistically significant depletion (stages III and IV vs. stages I and II) in only three classic MSGs (NM23 H2, ARHGDIB, and RECK), and one candidate MSG (PTPN11) [Figure 1]. Importantly, NM23 H2, a member of the Non Metastatic 23 (NM23) family showed very significant change in expression (P < 0.005). In order to ascertain the extent of deregulation, NM23 H2 expression was specifically examined in above clinical profiles; significant decrease in NM23 H2 was found in advanced stages [[Figure 1]; P < 0.005]. Decreased expression of NM23 H2 in metastatic NSCLC has been probed earlier, however, its prognostic significance has been unclear, and mechanisms of NM23 H2 action as a metastasis suppressor are poorly understood. A previous study of NM23 H2 in lung carcinoma derived A549 cells indicated its gene expression regulatory potential [27] suggesting a possible transcriptional role in progression of lung carcinoma. Though several studies have demonstrated NM23 H2 association with promoters, pointed out amino acids involved in interaction with DNA, and shown independence of regulatory function from enzymatic activity, [28] the precise contribution of NM23 H2 in suppression of metastasis has remained poorly understood. Notably, in our lab, NM23 H2 expression showed profound changes in transcriptome profile in lung cancer cells. Specifically, genes related to cell adhesion, epithelial organization and related signaling pathways were perturbed, supporting anti-metastatic action of NM23 H2. Since a causative role for NM23 H2 or the other MSG could lead to a new therapeutic approach in prevention of lung cancer metastasis, it is currently an active area of investigation.
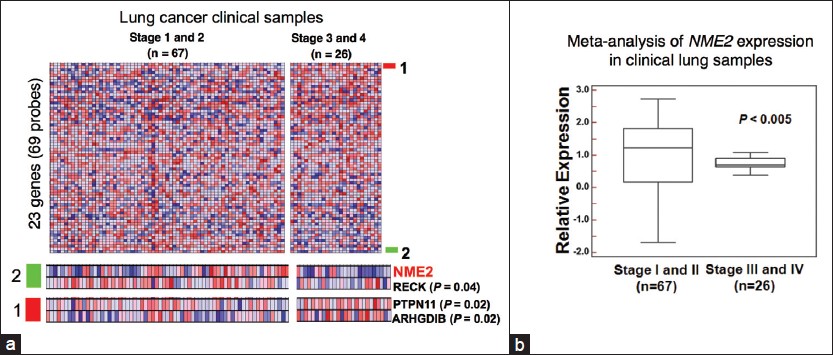 |
Figure 1: Expression analyses suggested NM23 H2 as a candidate biomarker metastasis suppressor gene in lung cancer. (a) Heat map representing transcript levels of 23 metastasis suppressor genes (MSG) (upper panel) in early/advanced lung cancer transcriptomes; 4 MSGs showed change in transcript level with NM23 H2 being most significant across tumors grouped stage wise (lower panel). (b) Box plot for relative expression of NM23 H2 in 93 lung cancer transcriptomes. Gene expression values were Z score normalized for comparison. Significance was calculated using student’s t-test Click here to view |
In the preceding sections, a brief overview of DNA and protein biomarkers of lung cancer was provided, with examples of how each aspect of lung cancer can have different sets of biomarkers, e.g., risk of cancer, presence of cancer, type of cancer, likelihood of metastasis, probability of response to targeted therapy, and prognosis. Additional molecular types (miRNAs, small molecules, voltile organic compounds) from a larger range of biological substrates (breath, breath condensate, urine, saliva) are being reported. [29] This explosion is attributable to advances in molecular strategies and analytical platforms, including genomics, epigenomics, proteomics, and metabolomics, but has not yet changed clinical management. Since the full fabric of this busy field cannot be captured in a mini-review, we have chosen to focus upon two important emerging threads, which may have a large clinical impact in coming years.
Exhaled Breath Biomarkers in Lung Cancer
Exhaled breath contains volatile organic compounds (VOC) from the environment and the lungs, as well as aerosolized airway lining fluid. An exhaled breath condensate (EBC) can be obtained by passing exhaled breath over a very cold surface. Linus Pauling first attempted characterization of VOC in human breath in 1971. [30] Subsequently, more than 3000 exogenous and endogenous VOC have been detected in normal human breath, contributed by environmental inhalation or by physiological end-products. [31] The rationale behind the study of VOC biomarkers for lung cancer is that in the cancerous state, altered metabolic and biochemical pathways may produce and process VOCs differently from normal cells. [32] The ease of use and high degree of lung-specificity make this an attractive proposition, if validated. [33] Promising data has now emerged from several gas chromatography/mass spectrometry (GC/MS) based studies that have reported such biomarkers, but unfortunately, none has reached clinical usability so far. [34],[35],[36],[37] Potential marker compounds are alcohols, aldehydes, ketones and hydrocarbons, some of which are differentially found in breath of normal subjects and cancer patients [34] Specific sensor array based methods for the detection of volatile biomarkers have also reached significant advancement. A recent study used solid-phase microextraction and GC/MS to identify 42 VOC that represent lung cancer biomarkers. Four of these were used to train and optimize the gold nanoparticle based sensors, demonstrating good agreement between patient and simulated breath samples, showing that a simple e-nose for cancer may not be far away. [35]
However, given the relative lack of clinical successes so far, an emerging direction in this field is the application of systems biology methods to biomarker datasets. This is a natural offshoot of the realization that no single molecule can sufficiently discriminate complex processes, without context. Recent studies that used statistical approaches incorporating patient data, such as smoking status, have found that different sets of VOC were discriminatory for cancer in non-smokers and smokers. [38] Such mathematical models not only aid in the identification of multi-component markers with high discriminatory power, but also may enhance mechanistic understanding of the disease mechanisms. However, the level of computational analysis depends on the experimental data in hand, which so far is not much.
We speculate that an ideal methodology of volatile-based biomarkers should include system biology approaches in addition to chemometrics [Figure 2]a. To illustrate this point, results of a pilot-scale study conducted in an author’s (Ranjan Nanda) lab are shown, where exhaled breath collected over packed polymer Tenax tubes was analyzed by GC/MS. We identified 140 VOCs present in the breath of 9 lung cancer and 18 healthy subjects. Multivariate analysis of this data was conducted using Partial Least Squares Discriminant Analysis (PLS-DA) to establish a preliminary classification model and evaluate the relation between compounds and classes. Of 140 metabolites that were identified, 18 were significantly altered in lung cancer [Figure 2]b, which can then be visualized as networks for studying interrelationships [Figure 2]c. The dataset shown is for illustrative purposes only, and larger studies are needed for reliable biomarker discovery.
 |
Figure 2: Breath volatile organic compounds could be useful for non-invasive diagnosis as well as enhance system-level understating of biochemical changes of lung cancer. (a) Schematic workflow. (b) Partial Least Squares Discriminant Analysis score plot showing grouping trends (Lung cancer [LC]) and Healthy (h). (c) The network of the selected metabolites (identified in the study) and genes that might be involved in LC Click here to view |
EBC is also a useful substrate for small-molecule biomarker discovery, although much less convenient. Nuclear Magnetic Resonance spectroscopy based metabolomic studies of EBC have been found to discriminate between healthy and asthmatic subjects. [39] A limitation of EBC is lack of effective data normalization strategies and currently complex statistical strategies are required for multi-parametric data.
Mirnas As Biomarkers for Diagnosis And Treatment of Lung Cancer
miRNAs are ultrashort (18-25 nucleotides), non-coding RNA molecules that can specifically bind to target mRNA sequences, usually resulting in mRNA degradation. They are highly stable, being relatively resistant to nucleases, and can be found in most bodily fluids. miRNA expression profiles are highly dynamic, yet regulated, which makes them very suitable as biomarkers. [40] However, normalization of miRNA expression is difficult because of the lack of a “housekeeping” stably expressed miRNA. [41] Bianchi et al., have reported that asymptomatic lung cancer detected by CT screening is also associated with changes in circulating miRNA profile. [42] A multivariate risk-predictor algorithm based on the weighted linear combination of the 34-miRNA expression levels, was sufficient to identify patients with early stage NSCLCs in a population of asymptomatic high-risk individuals with 80% accuracy. [43] This relatively simple test, requiring only 1 ml blood and quantitative-polymerase chain reaction (Q-PCR), shows the potential to discriminate between benign and malignant CT lesions. Independent validation of this important report is awaited. Tumor miRNA profiles have also been shown to predict recurrences of localized stage I non-small cell lung cancer after surgical resection. [44] Whether circulating miRNA profiles can be used instead, remains to be seen.
In summary, while much progress has been made in biomarker discovery for lung cancer, much more needs to be done. For early detection, tests using exhaled breath or circulating miRNA appear to be particularly promising, although both fields are still young. So far, prognostic and therapeutic markers rely upon isolation of cancerous tissue, whether from lung or from circulating tumor cells. While miRNAs appear to be promising, it is unlikely that VOCs would be useful in this regard. Unpublished work from our lab (Anurag Agrawal) shows that it is possible to measure miRNAs in exhaled breath condensate by Q-PCR.This has the potential to create simple non-invasive breath based tests, incorporating the best of all. The field of miRNA based biomarkers in lung cancer is starting to bloom and further investigations are warranted.
References
| 1. | Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127:2893-917.  |
| 2. | National Lung Screening Trial Research Team, Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409.  |
| 3. | Brambilla E, Gazzeri S, Lantuejoul S, Coll JL, Moro D, Negoescu A, et al. p53 mutant immunophenotype and deregulation of p53 transcription pathway (Bcl2, Bax, and Waf1) in precursor bronchial lesions of lung cancer. Clin Cancer Res 1998;4:1609-18.  |
| 4. | Niklinski J, Niklinska W, Chyczewski L, Becker HD, Pluygers E. Molecular genetic abnormalities in premalignant lung lesions: Biological and clinical implications. Eur J Cancer Prev 2001;10:213-26.  |
| 5. | Shiono S, Omoe K, Endo A. K-ras gene mutation in sputum samples containing atypical cells and adenocarcinoma cells in the lung. Carcinogenesis 1996;17:1683-6.  |
| 6. | Aviel-Ronen S, Blackhall FH, Shepherd FA, Tsao MS. K-ras mutations in non-small-cell lung carcinoma: A review. Clin Lung Cancer 2006;8:30-8.  |
| 7. | Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer 2004;4:707-17.  |
| 8. | Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res 1999;59:67-70.  |
| 9. | Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene 2002;21:6915-35.  |
| 10. | Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007;450:1235-9.  |
| 11. | Schiller JH. Noninvasive monitoring of tumors. N Engl J Med 2008;359:418-20.  |
| 12. | Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev 2009;28:369-78.  |
| 13. | Wang Q, Wang S, Wang H, Li P, Ma Z. MicroRNAs: Novel biomarkers for lung cancer diagnosis, prediction and treatment. Exp Biol Med (Maywood) 2012;237:227-35.  |
| 14. | Sung HJ, Cho JY. Biomarkers for the lung cancer diagnosis and their advances in proteomics. BMB Rep 2008;41:615-25.  |
| 15. | Vrabec BB, Gajovic S. Molecular biomarkers of lung carcinoma. Front Biosci (Elite Ed) 2012;4:865-75.  |
| 16. | Hida T, Ogawa S, Park JC, Park JY, Shimizu J, Horio Y, et al. Gefitinib for the treatment of non-small-cell lung cancer. Expert Rev Anticancer Ther 2009;9:17-35.  |
| 17. | Okano T, Kondo T, Fujii K, Nishimura T, Takano T, Ohe Y, et al. Proteomic signature corresponding to the response to gefitinib (Iressa, ZD1839), an epidermal growth factor receptor tyrosine kinase inhibitor in lung adenocarcinoma. Clin Cancer Res 2007;13:799-805.  |
| 18. | Postel-Vinay S, Vanhecke E, Olaussen KA, Lord CJ, Ashworth A, Soria JC. The potential of exploiting DNA-repair defects for optimizing lung cancer treatment. Nat Rev Clin Oncol 2012;9:144-55.  |
| 19. | Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: From discovery to therapy in record time. Cancer Cell 2010;18:548-51.  |
| 20. | Shaw AT, Solomon B. Targeting anaplastic lymphoma kinase in lung cancer. Clin Cancer Res 2011;17:2081-6.  |
| 21. | Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6.  |
| 22. | Gupta GP, Massagué J. Cancer metastasis: Building a framework. Cell 2006;127:679-95.  |
| 23. | Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, et al. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst 1988;80:200-4.  |
| 24. | Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, et al. KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995;268:884-6.  |
| 25. | Gildea JJ, Seraj MJ, Oxford G, Harding MA, Hampton GM, Moskaluk CA, et al. RhoGDI2 is an invasion and metastasis suppressor gene in human cancer. Cancer Res 2002;62:6418-23.  |
| 26. | Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, et al. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst 1996;88:1731-7.  |
| 27. | Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, et al. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res 2009;37:172-83.  |
| 28. | Postel EH, Berberich SJ, Rooney JW, Kaetzel DM. Human NM23/nucleoside diphosphate kinase regulates gene expression through DNA binding to nuclease-hypersensitive transcriptional elements. J Bioenerg Biomembr 2000;32:277-84.  |
| 29. | Hassanein M, Callison JC, Callaway-Lane C, Aldrich MC, Grogan EL, Massion PP. The state of molecular biomarkers for the early detection of lung cancer. Cancer Prev Res (Phila) 2012;5:992-1006.  |
| 30. | Pauling L, Robinson AB, Teranishi R, Cary P. Quantitative analysis of urine vapor and breath by gas-liquid partition chromatography. Proc Natl Acad Sci U S A 1971;68:2374-6.  |
| 31. | Phillips M, Herrera J, Krishnan S, Zain M, Greenberg J, Cataneo RN. Variation in volatile organic compounds in the breath of normal humans. J Chromatogr B Biomed Sci Appl 1999;729:75-88.  |
| 32. | Mazzone PJ. Analysis of volatile organic compounds in the exhaled breath for the diagnosis of lung cancer. J Thorac Oncol 2008;3:774-80.  |
| 33. | Amann A, Spanĕl P, Smith D. Breath analysis: The approach towards clinical applications. Mini Rev Med Chem 2007;7:115-29.  |
| 34. | Bajtarevic A, Ager C, Pienz M, Klieber M, Schwarz K, Ligor M, et al. Noninvasive detection of lung cancer by analysis of exhaled breath. BMC Cancer 2009;9:348.  |
| 35. | Peng G, Tisch U, Adams O, Hakim M, Shehada N, Broza YY, et al. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nat Nanotechnol 2009;4:669-73.  |
| 36. | Phillips M, Altorki N, Austin JH, Cameron RB, Cataneo RN, Greenberg J, et al. Prediction of lung cancer using volatile biomarkers in breath. Cancer Biomark 2007;3:95-109.  |
| 37. | Poli D, Carbognani P, Corradi M, Goldoni M, Acampa O, Balbi B, et al. Exhaled volatile organic compounds in patients with non-small cell lung cancer: Cross sectional and nested short-term follow-up study. Respir Res 2005;6:71.  |
| 38. | Ulanowska A, Kowalkowski T, Trawiñska E, Buszewski B. The application of statistical methods using VOCs to identify patients with lung cancer. J Breath Res 2011;5:046008.  |
| 39. | Sinha A, Krishnan V, Sethi T, Roy S, Ghosh B, Lodha R, et al. Metabolomic signatures in nuclear magnetic resonance spectra of exhaled breath condensate identify asthma. Eur Respir J 2012;39:500-2.  |
| 40. | Mallick R, Patnaik SK, Yendamuri S. MicroRNAs and lung cancer: Biology and applications in diagnosis and prognosis. J Carcinog 2010;9:8.  [PUBMED]  |
| 41. | Yendamuri S, Kratzke R. MicroRNA biomarkers in lung cancer: MiRacle or quagMiRe? Transl Res 2011;157:209-15.  |
| 42. | Bianchi F, Nicassio F, Veronesi G, di Fiore PP. Circulating microRNAs: Next-generation biomarkers for early lung cancer detection. Ecancermedicalscience 2012;6:246.  |
| 43. | Bianchi F, Nicassio F, Marzi M, Belloni E, Dall’olio V, Bernard L, et al. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol Med 2011;3:495-503.  |
| 44. | Patnaik SK, Kannisto E, Knudsen S, Yendamuri S. Evaluation of microRNA expression profiles that may predict recurrence of localized stage I non-small cell lung cancer after surgical resection. Cancer Res 2010;70:36-45.  |
Authors
Dr. Ram Krishna Thakur, CSIR Institute of Genomics and Integrative Biology Delhi, India
Dr. Vinod Kumar Yadav, CSIR Institute of Genomics and Integrative Biology Delhi, India
Dr. Ranjan Nanda, International Center for Genetic Engineering and Biotechnology, Delhi, India
Dr. Shantanu Chowdhury, CSIR Institute of Genomics and Integrative Biology Delhi, India
Ms. Sangeetha Subramaniam, International Center for Genetic Engineering and Biotechnology, Delhi, India
Dr. Anurag Agrawal, CSIR Institute of Genomics and Integrative Biology Delhi, India
Figures
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
