Nidhi Anand1, Nuzhat Husain1, Renu Varshney1, Kiran Preet Malhotra1, Mohammad Kaif2
1Department of Pathology, Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow, Uttar Pradesh, India
2Department of Neurosurgery, Dr. Ram Manohar Lohia Institute of Medical Sciences, Lucknow, Uttar Pradesh, India
| Date of Submission | 27-May-2021 |
| Date of Decision | 23-Jun-2021 |
| Date of Acceptance | 28-Jun-2021 |
| Date of Web Publication | 11-Oct-2021 |
Correspondence Address:
Nidhi Anand
Department of Pathology, Dr. Ram Manohar Lohia Institute of Medical Sciences, Vibhuti Khand, Gomti Nagar, Lucknow, Uttar Pradesh
India
Source of Support: None, Conflict of Interest: None
DOI: 10.4103/jcar.jcar_17_21
Abstract
BACKGROUND: Diffuse gliomas in the adult population are the most common primary central nervous system (CNS) tumors. The World Health Organization incorporated isocitrate dehydrogenase (IDH) mutations and 1p/19q co-deletion with histopathological features into an “integrated diagnosis” in the revised classification of tumors of CNS. These molecular subgroups of diffuse gliomas are found to stratify patients into prognostically distinct groups better than the histological classification. The objectives of the current study were to assess the frequency of IDH mutation, ATRX expression loss, p53 overexpression, and 1p/19q co-deletion detection in adult diffuse gliomas (Grade II, III, and IV) and to correlate them with clinicopathological and histopathological features.
MATERIALS AND METHODS: The current study was a tertiary care hospital-based retrospective case series of 112 cases of adult diffuse gliomas. Immunohistochemistry (IHC)-based molecular detection was performed for IDH-1, ATRX, and p53 and fluorescent in situ hybridization (FISH) was performed for 1p/19q co-deletion detection.
RESULTS: IDH-1 mutation was present in 30.4% (n = 34/112) cases, ATRX expression was lost in 18% (n = 19/104) cases, p53 was mutated in 39.3% (n = 42/107) cases and 1p19q was co-deleted in 25% (n = 4/16) cases. In the IDH1 mutant cases, with retained ATRX, FISH for 1p/19q co-deletion was performed and was co-deleted in four cases.
CONCLUSION: The results of the present study indicate that IHC including IDH1/2, ATRX, and p53 is useful for the molecular classification of diffuse gliomas, which could be useful for the evaluation of prognosis, especially Grade III and II. Although the immunohistochemical approach does not replace genetic testing completely, it is a practical and powerful means of assessing molecular genetic changes. IDH mutations are the established markers of better prognosis in diffuse gliomas.
Keywords: ATRX, diffuse glioma, isocitrate dehydrogenase-1, molecular classification.
| How to cite this article: Anand N, Husain N, Varshney R, Malhotra KP, Kaif M. Molecular classification and stratification of adult diffuse gliomas: A tertiary care center study. J Carcinog 2021;20:20 |
| How to cite this URL: Anand N, Husain N, Varshney R, Malhotra KP, Kaif M. Molecular classification and stratification of adult diffuse gliomas: A tertiary care center study. J Carcinog [serial online] 2021 [cited 2021 Oct 13];20:20. Available from: https://carcinogenesis.com/text.asp?2021/20/1/20/328121 |
Introduction
Diffuse gliomas in the adult population are the most common primary central nervous system (CNS) tumors.[1] The diagnosis of diffuse gliomas was primarily based on the histopathological features before the revised 4th edition of the classification of tumors of the CNS by the World Health Organization (WHO) in 2016. The WHO incorporated isocitrate dehydrogenase (IDH) mutations and 1p/19q co-deletion with histopathological features into an “integrated diagnosis” in the revised classification of tumors of CNS.[2] The revised classification more distinctly defines diffuse gliomas from astrocytomas that are more circumscribed, that lack IDH gene mutations and may contain BRAF mutations (namely pilocytic astrocytoma, pleomorphic xanthoastrocytoma, and sub-ependymal giant cell astrocytoma).[3] In the diffuse gliomas category, Grade II and III astrocytomas and oligodendrogliomas along with Grade IV glioblastomas (GBMs) are included. The new WHO categories of adult gliomas on the molecular basis are diffuse and anaplastic astrocytoma, IDH-mutant type or IDH-wild type (WT); GBM, IDH-mutant or IDH-WT; oligodendroglioma and anaplastic oligodendroglioma, 1p19q co-deleted and IDH-mutant and diffuse midline glioma H3K27M-mutant.[2] These molecular subgroups of diffuse gliomas are found to stratify patients into prognostically distinct groups better than the histological classification.
IDH is the most important molecular marker to categorize diffuse gliomas. Both IDH 1 and IDH 2 mutations are seen in infiltrating gliomas. IDH 1 is the most common mutations of the IDH mutations found in gliomas. The most frequent IDH 1 mutation is R132H at codon 132 with the exchange of amino acid arginine for histidine. IDH mutant gliomas have a slower progression over time and have a better prognosis in comparison to IDH WT counterparts irrespective of their histological grade. These IDH mutations are thought to be important initial events in glioma-genesis as indicated by the fact that they occur in both astrocytic and oligodendroglial gliomas.[4] IDH-WT GBMs account for 90% of all GBMs and are typically arise de novo, without a precursor lesion and known as primary GBMs. About 10% of all GBMs are IDH-mutant type and known as secondary GBMs that develop through a malignant progression from Grade II or III astrocytomas.[5] Secondary GBMs have a better prognosis than primary GBMs and are usually located in the frontal lobe and show early presentation. Among several molecular parameters, the key markers for the subtyping of diffuse gliomas include IDH mutation, 1p/19q co-deletion, and alpha-thalassemia/mental retardation syndrome X-linked (ATRX) gene mutation along with other molecular markers such as p53, TERT, and Ki-67.[3],[6] Immunohistochemistry (IHC) is the most prevalent and acceptable method worldwide to demonstrate IDH1 (R132H) mutant protein and showed 100% concurrence with sequencing for IDH1 (R132H).[7]
ATRX gene is involved in chromatin remodeling, histone regulation, nucleosome assembly, and the maintenance of telomeres. ATRX gene mutation, located at Xq210.1 results in the loss of function. IHC is a reliable method to detect ATRX mutation where a tumor with the mutation shows negative immunostaining for ATRX protein, and positivity is seen in native glial/microglial cells, neurons, inflammatory cells, and endothelial cells. ATRX mutations are strongly correlated with tumors of astrocytic phenotype, which harbor TP53 and IDH mutations.[8],[9]
Oligodendrogliomas are the type of diffuse gliomas, which lack ATRX mutations and are characterized by IDH mutation and 1p/19q co-deletion. In 1p/19q co-deletion, there is an unbalanced translocation between the short arm of chromosome 1 (1p) with the long arm of chromosome 19 (19q) and is seen in oligodendrogliomas.[10] This co-deletion is a specific molecular modification and considered essential for the diagnosis of oligodendroglioma. This co-deletion is associated with improved survival and also serves as a predictor of sensitivity to chemotherapy.[11] 1p/19q co-deletion has been evaluated by the polymerase chain reaction based on two major methods, fluorescent in situ hybridization (FISH) and by different next-generation sequencing platforms. However, the preferred method for evaluating 1p/19q co-deletion is the FISH technique. 1p/19q co-deletion is not detectable by IHC, it is mutually exclusive with ATRX and p53 mutation in IDH mutant gliomas.[10]
TP53 is a crucial regulator of the cell cycle, forming a part of the tumor-suppressor gene family. The TP53 gene provides directions for making a protein called tumor protein p53 (or p53) that is a tumor suppressor. TP53 mutations are found to be nearly mutually exclusive with 1p/19q co-deletion in gliomas and are strongly correlated with an astrocytic morphology.[12] TP53 mutations are present in 50% of astrocytomas, in contrast to just 10% of oligodendrogliomas. As ATRX mutations and IDH mutations show a close relationship to TP53 mutations, these three mutations can be co-existed as a molecular signature of astrocytomas.[13] Various studies showed that the frequency of p53 mutation is higher in high-grade gliomas and are associated with poor prognosis and overall survival.[14] On IHC, p53 immunostaining is detected as positive nuclear staining and serves as a surrogate marker for this mutation. However, the gold standard test to detect the presence of a TP53 mutation in a particular sample is DNA sequencing. In the current study, IHC-based molecular detection was performed for IDH-1, ATRX and p53 and FISH was performed for 1p/19q co-deletion detection.
Based on this background, the current study was conducted as an algorithm-based stepwise analysis. The objectives of the current study were to assess the frequency of IDH mutation, ATRX expression loss, p53 overexpression, and 1p/19q co-deletion detection in adult diffuse gliomas (Grade II, III, and IV) and to correlate them with clinicopathological and histopathological features.
Materials and Methods
Patient selection
The current study was a tertiary care hospital-based retrospective case series of 112 cases of adult diffuse gliomas that were categorized according to the WHO 2016 classification of the tumors of CNS. The study was approved by the Institutional Ethical Committee (IEC-14/15). Archived slides and paraffin blocks for all the cases with histological diagnosis of diffuse gliomas including diffuse astrocytomas, oligodendrogliomas, and GBMs in adult populations were obtained from the pathology department. Cases with poor processing and posttreatment were excluded. Relevant clinical and radiological details were obtained in all cases.
Histopathological evaluation
Histopathological evaluation was performed on hematoxylin and eosin stained archival slides from all the cases. The presence or absence of atypia, mitosis, and necrosis was assessed. The diagnosis of all the cases was confirmed and categorized according to the WHO 2016 classification of the tumors of CNS. Tumors showing only cytologic atypia along with variation in nuclear shape/size were graded as WHO Grade II and when anaplasia and unequivocal mitoses were also present, the tumors were labeled as Grade III. Tumors that additionally showing microvascular proliferation and/or necrosis were qualified as the WHO Grade IV.[3],[15]
Immunohistochemical evaluation
IHC for IDH1, ATRX and p53 has performed on the formalin-fixed paraffin-embedded tissue blocks according to the manufacturer’s protocols using the Ventana Benchmark XT system. The IHC for all the three markers were interpreted as per the standard guidelines for the markers.
Isocitrate dehydrogenase 1 immunostaining and interpretation
The IHC clone used was anti-IDH1 R132H (Dianova, USA Clone: H09-unconj) in dilution of 1:25. Immunoreactions were scored positive for mutated IDH1 protein when a majority of tumor cells showed strong cytoplasmic positivity with or without nuclear staining.[16] Normal/residual glial cells and vascular endothelial cells served as an internal negative control.
ATRX immunostaining and interpretation
The IHC clone used was Anti ATRX (HPA001906, Sigma, USA) in dilution of 1:100. >10% loss of nuclear expression in tumor cells was interpreted as positive mutation. Nonneoplastic cells such as endothelia, microglia, lymphocytes, and reactive astrocytes serve as a positive internal control.
p53 immunostaining and interpretation
The IHC clone used was p-53 protein (DO-07-Dakopatts, Denmark), Ready To Use (RTU). Intense staining of the nucleus in >30% of the tumor cells was considered a positive mutation. Positive controls and negative controls (by omitting primary antibody) were run with all batches.
Fluorescent in situ hybridization for 1p/19q co-deletion and interpretation
The cases which favored the diagnosis of oligodendroglioma on morphology and IHC, FISH for 1p/19q co-deletion were performed using the ZytoLight FISH Tissue Implementation Kit (which uses ZytoLight SPEC 1p36/1q25 Dual Color Probe and ZytoLight SPEC 19q13/19p13 Dual Color Probe). A control tissue cell lines (positive or negative) were run with every case.
Tissue microarray preparation
Representative areas from the tumor were marked on the slide and then on the corresponding block. For each case, 2–3 tumor tissue cores (1.5–2.0 mm diameter) from representative tumor areas were punched out and embedded into a new paraffin array block using an automated tissue microarray (TMA) (TMA Grandmaster, company, USA).
Fluorescent in situ hybridization analysis
FISH analysis was performed on deparaffinized 4 μm tissue sections using the ZytoLight FISH Tissue Implementation Kit. Sections of 3–4 μ thickness were cut and mounted on a chemically coated slide (Biocare, USA). This was followed by slides baking, removal of wax and dehydration. Pretreatment was done by Pretreatment Solution Citric (PT1), followed by protease digestion. The probe was applied, followed by denaturation and hybridization in Thermobrite (hybridizer). After washing, application of 4, 6-dia-amidino- 2-phenylindole was done for counterstaining. The signals were evaluated in the fluorescent microscope. A total of 50 cells were counted in each case, cell with deletion was reported when ≤1 orange signal and two green signals were visualized. The presence or absence of deletion was determined based on the 1p: 1q and 19q: 19p copy number ratios. The deletion was defined as a ratio less than or equal to 0.8. In normal cells, two orange and two green signals were visualized. Positive and negative controls are assessed before the case assessment. After interpretation of IDH1, ATRX, P53 immunostaining, and FISH for 1p/19q, gliomas were categorized into molecular subtypes according to the WHO 2016 classification of the tumors of CNS. Cases with mutated IDH, retained ATRX, and 1p19q co-deleted by FISH were designated as oligodendrogliomas. Cases with lost ATRX were classified as astrocytomas, IDH mutant, and IDH WT depending upon the IDH immunostaining.
Statistical analysis
The statistical analysis was performed using the Statistical Package for the Social Sciences software version 20. IBM Statistical Package for the Social Sciences (SPSS) software version 20, USA.
Results
The current WHO 2016 classification of diffuse gliomas is based on the histopathological features and molecular parameters in comparison to the previous categorization that was based on the light microscopic appearance of the tumors and its line of differentiation. We studied 112 cases of adult diffuse gliomas in the current study between June 2016 and July 2018.
The overall mean age of the cases was 43.4 years with patients ranging in age from 18 years to 78 years. In Grade II tumors, the median age of the cases was 33.2 years; in Grade III tumors, 40.5 years; and in Grade IV tumors, 48.9 years.
The male-to-female (M: F) ratio was 2.4:1. The exact site of biopsy was available in 77.6% (n = 87/112) of cases, of which 35.6% (n = 31/87) were from frontal lobe, 9.2% (n = 8/87) from parietal, 11.5% (n = 10/87) temporal, 10% (n = 9/87) frontoparietal, 8.4% (n = 7/87) temporo parietal, 6.9% (n = 6/87) frontotemporal, 2.3% (n = 2/87) occipital, 8.4% (n = 7/87) parietooccipital, 3% (n = 3/87) from thalamus and one case each from pineal gland, CP angle, as supratentorial mass and Sylvian lesion. The most common site was frontal lobe 35.6% (n = 31/87). In 22.3% (n = 25/112) cases, the exact location was not mentioned.
A maximum number of cases were categorized as Grade IV, in 52.7% (n = 59/112) cases followed by Grade III, in 25% (n = 28/112) cases and Grade II, in 22.3% (n = 25/112) cases [Figure 1].
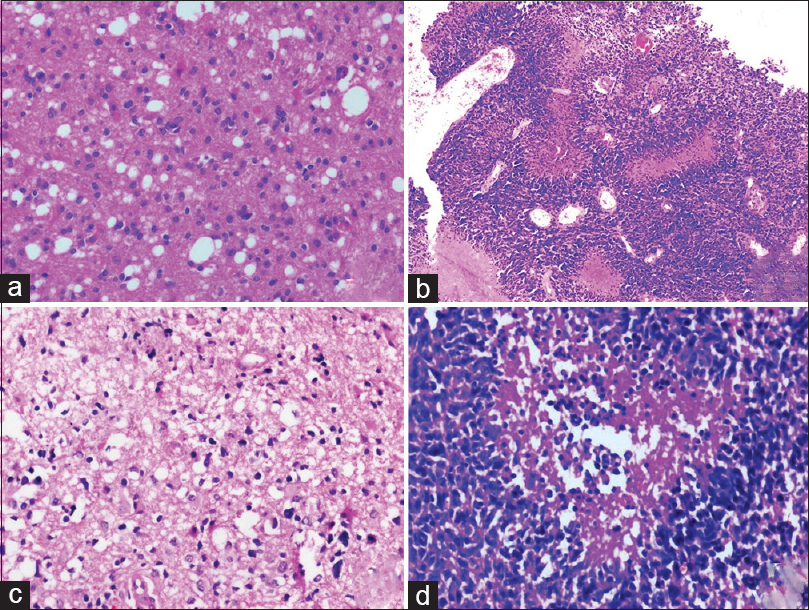 |
Figure 1: (a) Diffuse glioma World Health Organization Grade II (H and E, ×100), (b) Anaplastic astrocytoma World Health Organization Grade III (H and E, ×100), (c and d) Glioblastoma (C = H and E, ×50, d = H and E, ×200) Click here to view |
Overall, IDH-1 mutation was present in 30.4% (n = 34/112) cases, ATRX expression was lost in 18% (n = 19/104) cases, p53 was mutated in 39.3% (n = 42/107) cases, and 1p19q was co-deleted in 25% (n = 4/16) cases [Table 1] and [Figure 2].
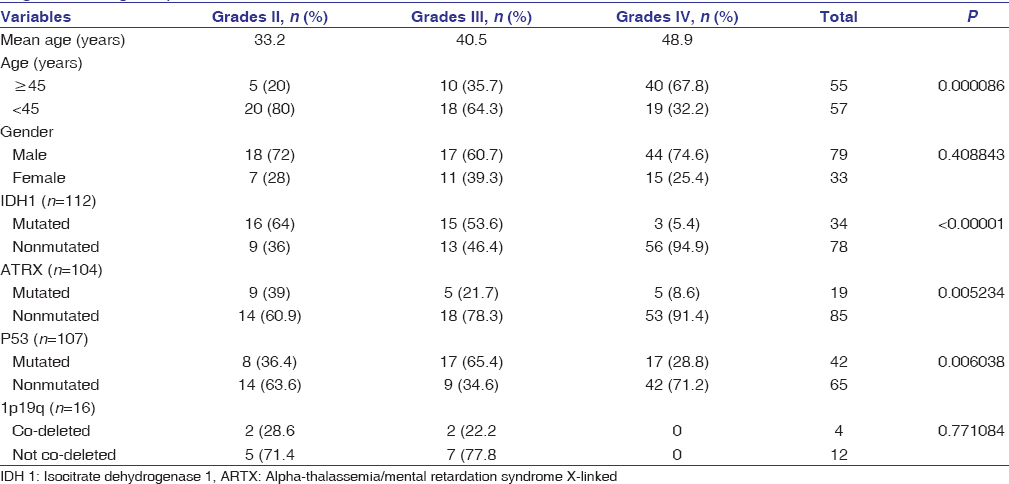 |
Table 1: Clinicopathological characteristics of the patients included the study (grouped by the World Health Organization grade) Click here to view |
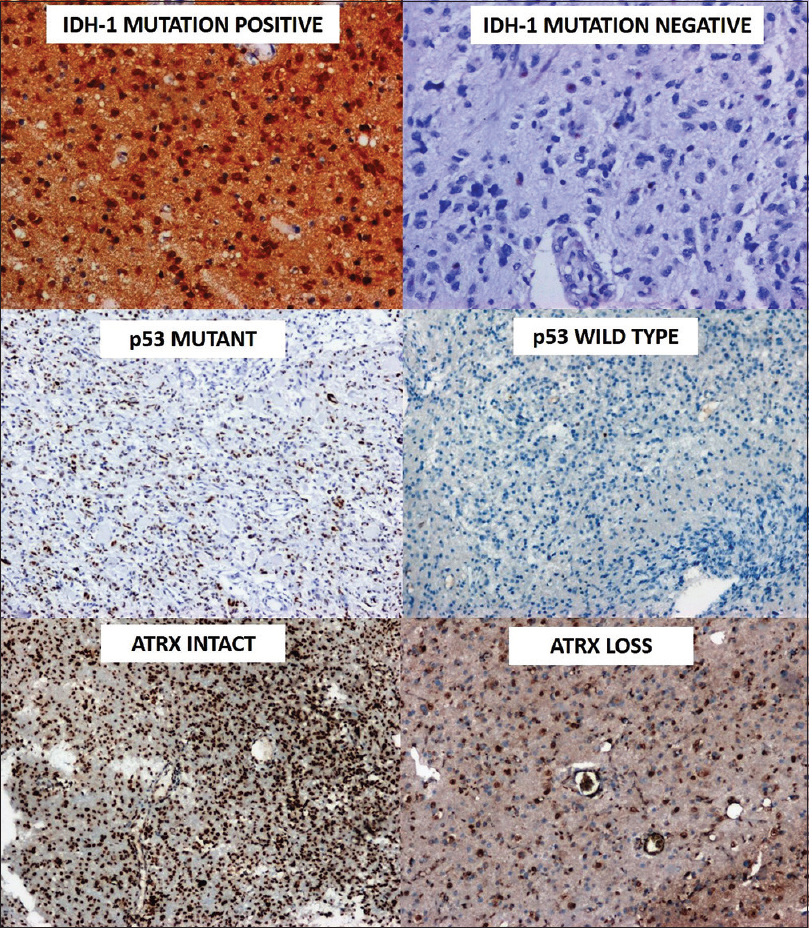 |
Figure 2: Immunohistochemistry for molecular testing demonstrating isocitrate dehydrogenase-1 expression, p-53 mutation and ATRX loss (3,3′-Diaminobenzidine (DAB), ×100) Click here to view |
Isocitrate dehydrogenase mutations
In total, 25 cases of Grade II lesions included in the study, IDH mutation was positive in 64% (n = 16/25) cases and negative in 36% (n = 9/25) cases. In 28 cases of Grade III lesions, 53.6% (15/28) cases were IDH1 mutated and 46.4% (n = 13/28) were IDH1 nonmutated. In Grade IV lesions, the total number of cases was 59, of which 5.1% (n = 3/59) were IDH1 mutation-positive and 94.9% (n = 56/59) were IDH1 mutation-negative. In IDH1-mutated cases, M: F ratio was 2.8:1, and the mean age was 40.3 years.
ATRX loss
In Grade II lesions, ATRX was lost in 39.1% (n = 9/23) cases and retained in 60.9% (n = 14/23) cases. In Grade III lesions, ATRX was lost in 21.7% (n = 5/23) cases and in 78.2% (n = 18/23) cases ATRX was intact. In Grade IV lesions, ATRX was lost in 8.6% (n = 5/58) cases, and in 91.4% (n = 53/58) cases, ATRX was intact. In ATRX mutated cases, M: F ratio was 8.5:1, and the mean age was 33.6 years.
Expression of p53
In Grade II lesions, p53 was mutated in 36.4% (n = 8/22) cases and WT/nonmutated in 63.6% (n = 14/22) cases; in Grade III lesions, it was mutated in 65.4% (n = 17/26) cases and WT/nonmutated in 34.6% (n = 9/26) cases; in Grade IV lesion, p53 was mutated in 28.8% (n = 17/59) cases and WT/nonmutated in 71.2% (n = 42/59) cases. In p53 mutated cases, M: F ratio was 2.8:1, and the mean age was 38.6 years.
1p19q co-deletion
Out of 20 cases of oligodendroglioma phenotype, FISH for 1p19q was performed in 16 cases. It was co-deleted in 28.6% (n = 2/7) cases of Grade II and 22.2% (n = 2/9) cases of Grade III lesions.
Discussion
The updated WHO 2016 classification of CNS tumors incorporated molecular classification with histology to provide an integrated diagnosis. For diffuse gliomas, mutations in IDH, ATRX, and 1p/19q co-deletion status are now crucial in the pathological diagnosis of glioma.[3] IDH mutation is one of the most important molecular markers in glioma.[2] The proposed molecular classifications of gliomas in the literature commonly include some combination of IDH, 1p/19q co-deletion and the tumor’s telomere maintenance mechanism, defined by alterations in either TERT or ATRX.[15]
The current study aimed to classify the adult diffuse gliomas after incorporating these molecular markers in the diagnosis and assess the frequency of completely classifiable cases. Incorporation of molecular classification of diffuse gliomas as a part of layered reporting in histopathology is essential in the regular clinical practice. The overall survival and treatment are significantly associated with the molecular stratification of tumors.
In the current study, the mean age of the patients was 43.4 years, and the M: F ratio was 2.4:1. In the current study, the most common site for the occurrence of diffuse glioma was the frontal lobe followed by the temporal and frontoparietal lobe. This finding is in concordance with the results of the study conducted by Javed et al.[3]
In the current study, IDH-1 mutations were identified in 30.4% of cases. In the study conducted by Malueka et al., which included 106 cases of gliomas, IDH mutations were identified in 21.7% of cases.[17] In the current study, IDH-1 mutations were identified in IDH mutation 64% Grade II tumors and only 5.1% of the Grade IV tumors. In the current study, this finding statistically correlated with the degree of anaplasia in the tumor. This finding is contrary to the results obtained from the study conducted by Malueka et al.[17] Wherein the authors inferred that about 40% of Grade IV tumors harbored the IDH-1 mutations. In the current study, 94.9% of the Grade IV tumors were IDH-1 negative. IDH is the key gene associated with the development and proliferation of gliomas. IDH WT tumors are believed to have an aggressive clinical course.[2],[4]
In the current study, ATRX loss was identified in 18% of cases, whereas the p53 mutations were detected in 39.3% of cases. In the study conducted by Nguyen et al., wherein the authors performed immunohistochemical analysis using TMAs and demonstrated that partial ATRX loss was not infrequent in diffuse gliomas. Tumors with p53 mutations are associated with a poor prognosis. The overall survival and disease-free survival were low among this group.[18],[19] In this study, mutations in IDH-1 and ATRX are not associated with certain histologic subgroups and grade of gliomas. The present study revealed the frequency of ATRX loss; it occurred frequently in the Grade II and III gliomas. These results are similar to those obtained from the study conducted by Wiestler et al. ATRX loss refines the classification of anaplastic gliomas and identifies a subgroup of IDH mutant astrocytic tumors with better prognosis.[20]
In the IDH1 mutant cases, with retained ATRX, FISH for 1p/19q co-deletion was performed and was co-deleted in four cases. They were classified as diffuse gliomas (oligodendroglioma phenotype), IDH mutant, 1p/19q co-deleted. Upon review of the molecular and histologic features, we were able to classify nearly all the study cases according to the new WHO 2016 criteria.
In the current study, the testing for all the molecular markers was done using the IHC. There are studies published in the literature that state IHC can be used as a surrogate marker for the molecular techniques.[5],[7] However, it is essential to ensure that procedural shortcomings are accurately rectified and controls (both positive and negative) are run with every batch.[21] In the current study, controls were run with every batch additionally staining results were evaluated in areas where internal controls were properly stained.
The results of the present study indicate that IHC, including IDH1/2, ATRX, and p53 is useful for the molecular classification of diffuse gliomas, which could be useful for the evaluation of prognosis, especially Grade III and II. Although the immunohistochemical approach does not replace genetic testing completely, it is a practical and powerful means of assessing molecular genetic changes. IDH mutations are the established markers of better prognosis in diffuse gliomas.
The limitation of this study is the lack of availability of treatment data and patient survival.
Conclusion
In summary, this study combines the cases of adult glioma patients that have been successfully re-classified according to the new integrated diagnosis criteria in the revised 4th edition of the WHO 2016.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
| 1. | |
| 2. | |
| 3. | |
| 4. | |
| 5. | |
| 6. | |
| 7. | |
| 8. | |
| 9. | |
| 10. | |
| 11. | |
| 12. | |
| 13. | |
| 14. | |
| 15. | |
| 16. | |
| 17. | |
| 18. | |
| 19. | |
| 20. | |
| 21. |
Figures
[Figure 1], [Figure 2]
Tables
[Table 1]

