Rifat Hasina1, Mosmi Surati1, Ichiro Kawada1, Qudsia Arif2, George B Carey1, Rajani Kanteti1, Aliya N Husain2, Mark K Ferguson3, Everett E Vokes1, Victoria M Villaflor1, Ravi Salgia1
1 Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL, USA
2 Department of Pathology, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL, USA
3 Department of Surgery, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL, USA
| Date of Submission | 16-Jul-2013 |
| Date of Acceptance | 06-Aug-2013 |
| Date of Web Publication | 28-Oct-2013 |
Correspondence Address:
Ravi Salgia
Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
USA
Source of Support: None, Conflict of Interest: None
DOI: 10.4103/1477-3163.120632
Abstract
Background: World-wide, esophageal cancer is a growing epidemic and patients frequently present with advanced disease that is surgically inoperable. Hence, chemotherapy is the predominate treatment. Cytotoxic platinum compounds are mostly used, but their efficacy is only moderate. Newer alkylating agents have shown promise in other tumor types, but little is known about their utility in esophageal cancer. Methods: We utilized archived human esophageal cancer samples and esophageal cancer cell lines to evaluate O-6-methylguanine-deoxyribonucleic acid methyltransferase (MGMT) hypermethylation status and determined sensitivity to the alkylating drug temozolomide (TMZ). Immunoblot analysis was performed to determine MGMT protein expression in cell lines. To assess and confirm the effect of TMZ treatment in a methylated esophageal cancer cell line in vivo, a mouse flank xenograft tumor model was utilized. Results: Nearly 71% (12/17) of adenocarcinoma and 38% (3/8) of squamous cell carcinoma (SCC) patient samples were MGMT hypermethylated. Out of four adenocarcinoma and nine SCC cell lines tested, one of each histology was hypermethylated. Immunoblot analyses confirmed that hypermethylated cell lines did not express the MGMT protein. In vitro cell viability assays showed the methylated Kyse-140 and FLO cells to be sensitive to TMZ at an IC 50 of 52-420 μM, whereas unmethylated cells Kyse-410 and SKGT-4 did not respond. In an in vivo xenograft tumor model with Kyse-140 cells, which are MGMT hypermethylated, TMZ treatment abrogated tumor growth by more than 60%. Conclusion: MGMT methylation may be an important biomarker in subsets of esophageal cancers and targeting by TMZ may be utilized to successfully treat these patients.
Keywords: Alkylating agents, deoxyribonucleic acid repair genes, in vivo pre-clinical, esophageal cancer, O-6-methylguanine-deoxyribonucleic acid methyltransferase hypermethylation, response to treatment, temozolomide
How to cite this article:
Hasina R, Surati M, Kawada I, Arif Q, Carey GB, Kanteti R, Husain AN, Ferguson MK, Vokes EE, Villaflor VM, Salgia R. O-6-methylguanine-deoxyribonucleic acid methyltransferase methylation enhances response to temozolomide treatment in esophageal cancer. J Carcinog 2013;12:20
How to cite this URL:
Hasina R, Surati M, Kawada I, Arif Q, Carey GB, Kanteti R, Husain AN, Ferguson MK, Vokes EE, Villaflor VM, Salgia R. O-6-methylguanine-deoxyribonucleic acid methyltransferase methylation enhances response to temozolomide treatment in esophageal cancer. J Carcinog [serial online] 2013 [cited 2021 Oct 13];12:20. Available from: https://carcinogenesis.com/text.asp?2013/12/1/20/120632
Background
Esophageal cancer is a growing epidemic with approximately 482,300 new esophageal cancer cases and 406,800 deaths world-wide. [1],[2] In the Western world, adenocarcinoma has increased in incidence while in East Asia and developing countries squamous cell carcinoma (SCC) remains problematic. The distinct histologies of adenocarcinoma of the gastro-esophageal junction and SCC of the esophagus are often classified and treated similarly, but have different risk factors and clinical courses. In patients who have locally advanced esophageal cancer, multimodality treatment is most common, whereas patients with late stage disease are treated palliatively with chemotherapy and occasionally radiation. Despite the most aggressive therapy, the relative 5 year survival for esophageal cancer is dismal. [3],[4] In order to make an impact on this disease, new biologic targets need to be determined.
Various biologic targets have been shown to be expressed and amplified in esophageal cancers. [5],[6],[7] Multiple attempts to inhibit epidermal growth factor receptor and vascular endothelial growth factor (VEGF) have not resulted in additional survival benefit. [7],[8],[9],[10],[11] Human epidermal growth factor receptor 2 (HER2/neu) has been noted to be amplified in a subset of esophageal and gastro-esophageal tumors and therapeutic inhibition with trastuzumab has been shown to be effective in HER2/neu overexpressed tumors by Bang et al. [12] However, the response was short-lived in a metastatic setting. MET receptor tyrosine kinase (RTK) has been shown to be expressed and sometimes amplified in esophageal tumors. [13] In a clinical trial evaluating crizotinib, responses were seen for MET-amplified esophageal cancers albeit for a short period of time. [13] Finally, overexpression of EPHB4 RTK and its potential role as a therapeutic target has been reported in esophageal cancers. [14] Even though a large number of laboratories are investigating the role of RTKs in esophageal cancers, epigenetic changes can occur in preneoplastic and neoplastic lesions, particularly, the hypermethylation of O-6-methylguanine-deoxyribonucleic acid (DNA) methyltransferase (MGMT). [15],[16],[17]
The MGMT gene encodes a DNA repair protein that removes alkyl groups from the 0-6 position of guanine, an important site of DNA alkylation. This methylation results in the epigenetic silencing of the MGMT gene. Studies, particularly in the field of brain tumor and melanoma research, have demonstrated that tumors with MGMT methylation have shorter progression free survival times. [18] However, such genetic silencing is also associated with improved overall survival in patients receiving alkylating agents. [19] Temozolomide (TMZ), an oral cytotoxic alkylating agent, is one of a new class of compounds known as imidazotetrazines. Alkylating chemotherapeutic agents bind to and damage DNA, thereby killing tumor cells. With diminished DNA-repair activity, MGMT methylated cancers have increased sensitivity to such therapy. Given that MGMT hypermethylation has been demonstrated in esophageal pre-malignant and malignant lesions, [20] we investigated the role of TMZ in the treatment of esophageal cancer.
Our recent experience with a patient with esophageal cancer whose tumor expressed methylated MGMT and had a durable response to TMZ stimulated us to explore the relative frequency of MGMT methylation in esophageal tumors. We also sought to determine whether MGMT methylated cell lines respond to TMZ inhibition, which could suggest a potential role for this pathway in the treatment of esophageal cancer.
Methods
Compliance with ethics guidelines
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
For studies with animals: All institutional and national guidelines for the care and use of laboratory animals were followed.
Cell lines
Esophageal cancer cell lines (BE-3, FLO, SKGT-4, SKGT-5, Kyse-110, Kyse-140, Kyse-220, Kyse-410, Kyse-520, Kyse-850, TE-1, TE-8 and TE-12) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) or DMEM+Hank’s F12 media supplemented with 10% fetal bovine serum and penicillin in a 37°C, 5% CO 2 environment. The Kyse cells were acquired through a generous gift from Dr. Yutaka Shimada of Toyama University, Japan, and the FLO cells were a generous gift from Dr. David D. Beers of University of Michigan, MI.
Tissue acquisition
Samples for this study were acquired from paraffin-embedded, formalin-fixed tissues archived at the Human Tissue Resource Center available to us through Institutional Review Board -approved protocols. An experienced Pathologist analyzed all samples using conventional light microscopy and determined the histological subtype as well as tumor grade. These data were entered into our HIPAA compliant database, which included demographic and clinical information as well as treatment outcomes.
Genomic DNA
gDNA was collected from esophageal adenocarcinoma and SCC samples available in the above group. DNA was purified according to procedures described previously. [21] Briefly, five sections at 10 μM thickness were lysed with digestion buffer and proteinase K at 56°C for 48 h. The DNA was chelated with Chelex-100 and the extracted supernatant was collected after centrifugation. The integrity of gDNA was evaluated by agarose gel electrophoresis followed by polymerase chain reaction (PCR) with beta globin. The gDNA was quantified using Nanodrop and stored at −20°C.
MGMT methylation detection
MGMT promoter methylation status was determined by methylation specific PCR performed on the above-described gDNA at the laboratories of LabCorp, Research Triangle Park, NC. Results were reported as methylated, unmethylated or undetected, which may have been due to poor quality or low DNA content of samples.
Viability assay
Cell viability assay was carried out using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, cells were seeded at a density of 4000 cells/well in 96 well plates and incubated overnight. Following this, the media was replaced with fresh media supplemented with TMZ at a final concentration of 1500 μM as the highest dose and eight fold serial dilutions of it. Dimethylsulfoxide (DMSO) was used as vehicle control. The viability of cells was assessed at 72 h of drug treatment. MTT reagent was added to each plate at the end point and cells incubated for 4 h, after which MTT solubilizing agent was added. Plates were read at 570 nm using Synergy HT multi-mode microplate reader. Percentage cell viability was calculated by comparing treated cells with DMSO control. The IC 50 of TMZ was also determined from this data.
Immunoblot
Cells were washed twice with ice-cold phosphate buffer solution and lysed using radio immunoprecipitation assay (RIPA) buffer (Boston Bioproducts, #BP115) to extract total protein. The lysis buffer was supplemented with sodium orthovanadate, protease inhibitors, and phosphatase inhibitor. Lysates were incubated with RIPA buffer for 45 min with vortexing every 10 min. 150 μg of total protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on 7.5% gel at 100 V, transferred to polyvinylidene difluoride membranes over 1 h at 22 V and probed with anti-human MGMT antibody (Millipore, MAB16200). Membranes were blocked with 5% bovine serum albumin at room temperature for 1 h. Protein bands were visualized using a horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz, SC-2031), exposed to 1:1 HRP ECL solutions (Bio-Rad, #170-5070) for 1 min and images taken using chemiluminescent imaging system ChemiDoc™ XRS (Bio-Rad). The ovarian cancer cell line A2780 was used as a positive control and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was probed to serve as a protein loading control.
In vivo xenograft tumor inhibition
2.5 million Kyse-140 cells were mixed 1:1 with Matrigel (BD Biosciences) and injected subcutaneously in the right flank of 6-week-old female nude mice (Harlan Laboratories). Mice were randomized into two groups of eight mice each when tumor volume reached an average of 250 mm 3 . One group received TMZ at 80 mg/kg administered intraperitoneally on days 6, 7, 8 and 9. The other group received vehicle control by same route and dosing schedule. Mice were allowed to grow freely and tumor volume measured every 3 rd day. The tumor volume was calculated by the formula: (height × width × length)/2 and plotted to compare the effect of drug treatment. On day 32, the vehicle control mice were euthanized due to high tumor volume. The TMZ treated mice were euthanized on day 50. After sacrifice, the tumors were harvested and submitted for histologic evaluation. All procedures were approved by Institutional Animal Care and Use Committee and performed in accordance with the Animal Welfare Act regulation.
Results
Case report
Herein we report a case of a 72-year-old man who had experienced 4-month history of dysphagia with solids and a subsequent 20 pound weight loss. An upper endoscopy revealed a partially obstructing mass located at 33 cm from the incisors, pathology characterized this tumor as moderately differentiated adenocarcinoma of the esophagus. A computed tomography (CT), endoscopic ultrasound and positron emission tomography (PET) were done as part of the staging work-up and the tumor was staged as T4N0M1. The PET scan revealed liver and adrenal metastasis [Figure 1]a. The patient received a regimen comprised of epirubicin, cisplatin and 5-fluorouracil (ECF). Patient tolerated ECF therapy fairly well with resolution of his dysphagia; however, he developed lower extremity edema and distal neuropathy, both of which partially resolved once the chemotherapy course was completed. On restaging, he was noted to have a mixed response with progressive liver disease following 2 cycles of chemotherapy by CT and PET scan. He tested positive for MGMT methylation and received TMZ.
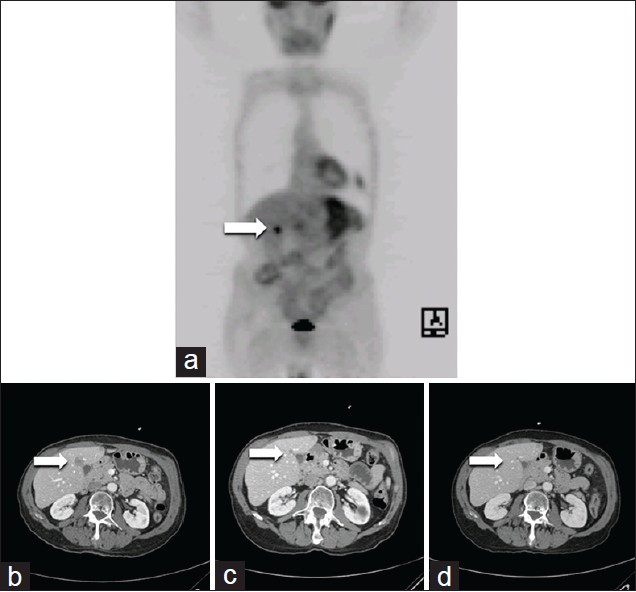 |
Figure 1: Response to TMZ in a patient with MGMT methylation. The arrow illustrates an area of increased uptake in the liver consistent with a hepatic metastasis on positron emission tomographic imaging. (a) This corresponds to the liver metastasis seen on the pre-treatment screening computed tomography scan. (b) Repeat CT imaging following 8 weeks (2 cycles) of treatment with temozolomide (TMZ) demonstrates marked reduction of hepatic metastasis. (c) Following 6 months of TMZ treatment, (d) Demonstrates complete resolution of the hepatic metastasis Click here to view |
The study protocol was to evaluate TMZ, an agent commonly used with brain tumors and melanoma. This phase 2 study, IRB approved, targeted individuals with advanced aerodigestive tract cancers whose tumors display methylation of their MGMT promoters. TMZ was given at 225 mg by mouth daily for 7 days followed by 7 days off the therapy with each cycle lasting 28 days. Oncology clinic visits with laboratory testing occurred monthly and scans for staging purposes occurred every 8 weeks. The TMZ was to continue until disease progression occurred or the side-effects of the treatment became intolerable.
Case result
Patient was treated with TMZ on protocol for a period of 18 months with continued response/stability of disease. Following the initial two cycles (2 months) of treatment with TMZ, a marked response was noted and at 6 months, a complete resolution of disease occurred [Figure 1]a-d. Treatment was tolerated for several cycles, but patient developed mild shortness of breath and was ultimately diagnosed with a bronchial infection and chronic obstructive pulmonary disease exacerbation. The TMZ was held briefly, the symptoms abated and chemotherapy was resumed. In addition, 6 months into therapy, the patient experiencing distal bilateral upper and lower extremity numbness and increasing difficulty with ambulation. An electromyography performed demonstrated a polyradiculoneuropathy with mixed axonal demyelinating features, consistent with chronic inflammatory demyelinating polyneuropathy (CIDP). CIDP was treated with intravenous immunoglobulin with improvements in gait and sensation.
An esophagogastroduodenoscopy done for dysphagia was notable for a severe 4 cm × 6 mm stenosis, which was stented. Subsequent to the stenting, the patient was found to have severe anemia likely secondary to slow bleeding from the site of the esophageal stent/tumor. Palliative radiotherapy of his local disease was given to control the gastrointestinal bleed. Patient died of progressive disease 24 months following treatment initiation.
MGMT methylation status in archival patient samples and cell lines
To further study MGMT methylation, 25 individual esophageal cancer patients’ tumor samples were evaluated [Table 1]. Due to the quality of archival tissues, many other samples were unsuitable for MGMT PCR assays. Of the 25 patient samples, 17 were adenocarcinoma and eight were SCC. The MGMT methylation status is summarized as: for adenocarcinoma, 12/17 (70.5%) cases were methylated and the remaining 5/17 (29.4%) cases were unmethylated. In addition, four adjacent normal tissue samples from the adenocarcinoma-methylated group were also found to be methylated. In SCC, 3/8 (37.5%) samples were methylated and 5/8 (62.5%) were unmethylated [Table 2]. In addition to patient tumor tissue, we also analyzed the MGMT methylation status of four esophageal adenocarcinoma and nine SCC cell lines. Of these, 1/4 (25%) adenocarcinoma cells were methylated and 1/9 (11%) SCC cells were methylated. The two hypermethylated cells were: FLO (adenocarcinoma) and Kyse-140 (SCC) [Table 2].
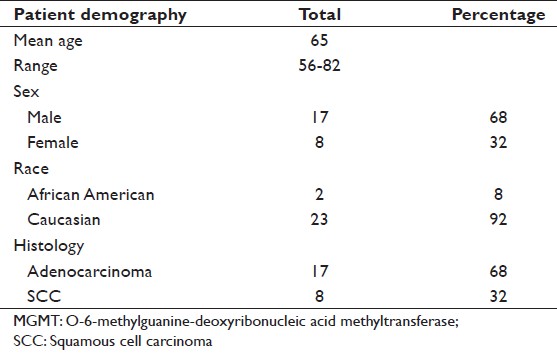 |
Table 1: Patient demographics of those whose MGMT methylation status was determined. In addition to esophageal carcinoma patient samples, four adenocarcinoma and nine SCC cell lines were also tested Click here to view |
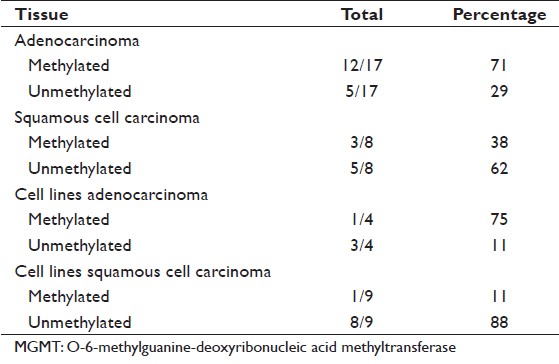 |
Table 2: The incidence of MGMT hypermethylation in tissues and cell lines tested Click here to view |
TMZ sensitivity in vitro
In order to determine the sensitivity of MGMT methylated esophageal cancer cell lines to TMZ treatment, methylated and unmethylated cell lines were subjected to TMZ treatment at different drug concentrations and their viability measured after 72 h. A methylated adenocarcinoma (FLO) and unmethylated adenocarcinoma SKGT-4 as well as a methylated SCC (Kyse-140) and unmethylated SCC Kyse-410 were used. In both histologies, the methylated cells (FLO, Kyse-140) showed increasing sensitivity to higher doses of TMZ (IC 50 52 μM and 420 μM respectively), whereas the unmethylated cells showed none or little sensitivity to the drug at the highest dose of 1500 μM [Figure 2].
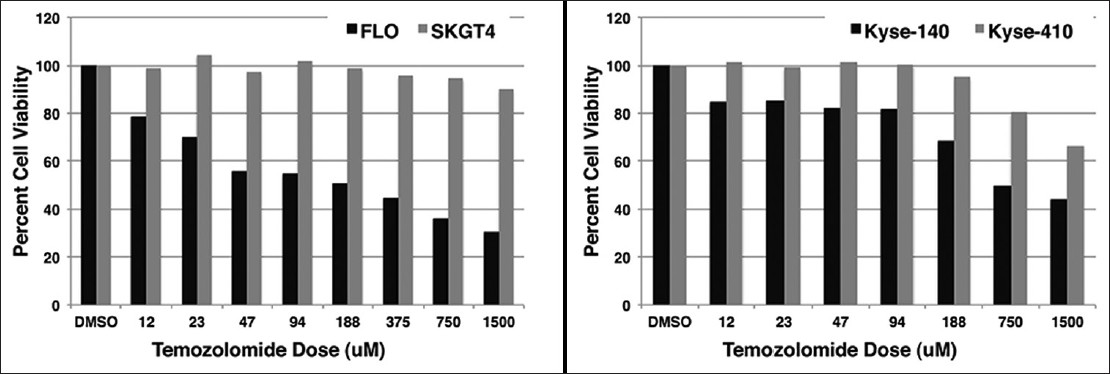 |
Figure 2: Response to TMZ in vitro. Methylated and unmethylated cells were treated with temozolomide (TMZ) for 72 h at different doses and their viability assessed. In both adenocarcinoma and squamous cell carcinoma, the methylated cells (FLO, Kyse-140) were sensitive to TMZ (IC50 52 μ M and 420 μ M respectively). The unmethylated cells (SKGT-4 and Kyse-410) showed little or no sensitivity to TMZ treatment Click here to view |
MGMT protein expression in oesophageal cancer cell lines
We analyzed the total MGMT protein expression in the above four esophageal cancer cell lines by immunoblotting with a specific antibody directed against MGMT. MGMT is a 22 kDa protein present in all normal tissues. In the two unmethylated cells (SKGT-4 and Kyse-410) there was robust expression of MGMT protein, whereas the methylated cell lines (FLO and Kyse-140) lost expression. A2780, an ovarian cancer cell line well-known to express MGMT was used as a positive control and GAPDH as a loading control [Figure 3].
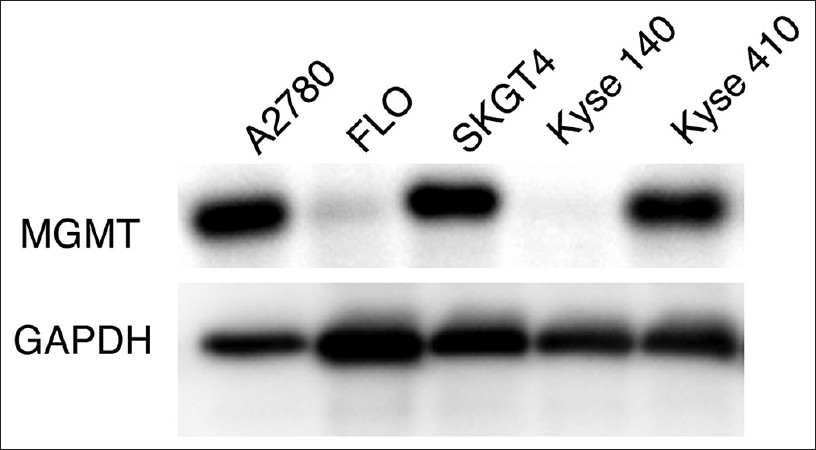 |
Figure 3: Expression of MGMT in esophageal cell lines. Methylated and unmethylated esophageal cancers cells were immunoblotted with anti-human O-6-methylguanine-deoxyribonucleic acid methyltransferase (MGMT) antibody. The data shows that total MGMT protein expression was lost in the two methylated cells. The ovarian cancer cell line A2780 was used as a positive control and glyceraldehyde-3-phosphate dehydrogenase was probed to serve as a protein loading control Click here to view |
TMZ sensitivity in an in vivo mouse model
To further test the sensitivity of methylated cells to TMZ treatment, a subcutaneous flank xenograft mouse model was used. The MGMT methylated SCC cell line Kyse-140 cells were subcutaneously implanted in the right leg and the mice treated with TMZ according to regimen detailed in materials and methods. In the vehicle control group, two out of the eight mice were terminated early on days 15 and 22 due to tumor volume exceeding allowed size. The remaining six mice were euthanized on day 32 [Figure 4]a and c. The TMZ treated mice had slower growing tumors and were allowed to live for 3 weeks after treatment ended until day 50 at which time they were euthanized [Figure 4]b and d. In this group also, two mice were terminated early on days 13 and 19 due to body weight loss. The tumor volumes of the two groups are plotted and shown in [Figure 4]e. The average tumor volume for the untreated group was 1708 mm 3 on day 32, when they were sacrificed. The average volume of the TMZ group was 662 mm 3 on day 32 and 1332 on day 50. The results show that there was significant inhibition of tumor growth by TMZ treatment compared with untreated control [Figure 4]e.
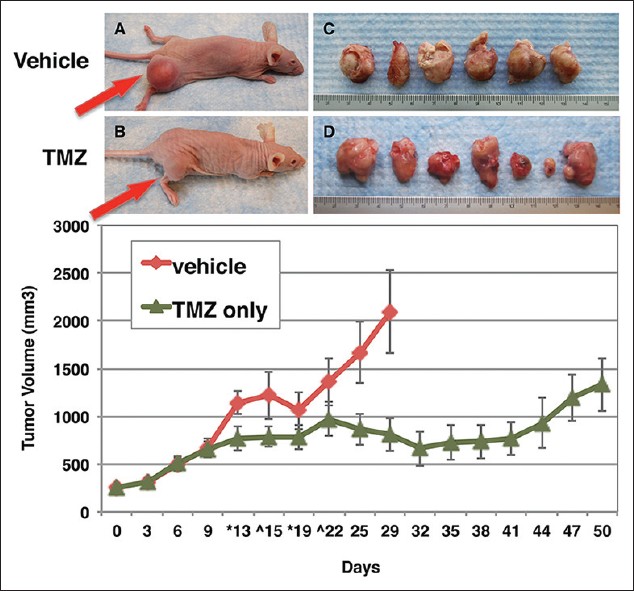 |
Figure 4: Response to TMZ in vivo. Mice bearing Kyse-140 cells were treated with temozolomide (TMZ) 80 mg/kg on days 6, 7, 8 and 9. (a) Representative vehicle mouse on day 32, at which time the group was sacrificed. (b) Representative mouse from TMZ treatment group on day 50, at which time this group was sacrificed. (c and d) Subcutaneously implanted leg tumors harvested from vehicle and TMZ treated groups respectively. (e) Tumor volume of treated and untreated groups (n = 6/8) plotted against time Click here to view |
Discussion
The clinical response to TMZ in a patient with MGMT methylated esophageal cancer led us to evaluate the role of MGMT methylation and inhibition. In an analysis of archived tissues samples, 71% of esophageal adenocarcinoma and 38% of SCC patient samples were positive for MGMT methylation. In in vitro, methylated esophageal cancer cells were sensitive to TMZ-induced cytotoxicity, whereas unmethylated cells displayed a lack of sensitivity to TMZ. The data presented in the manuscript shows the effects of TMZ on various cell lines without any genetic or pharmacological effects. This is a strong finding in vitro as well as in vivo. This helps us translationally to begin to understand our dramatic esophageal cancer case study. We showed that Kyse-140 also responded very strongly to TMZ in vitro at lower doses and compared it to another cell line of similar histology, Kyse-410, which had muted response even at the highest dose. These are standard methods of comparing drug effects among various cell lines.
In the era of targeted therapies, esophageal cancer is still under-investigated. We believe that MGMT methylation and targeting by TMZ is important in certain subsets of esophageal tumors. The majority of evidence for TMZ utilization comes from glioblastoma multiforme (GBM). [22],[23],[24] Patients with GBM containing a methylated MGMT promoter benefited from TMZ, whereas those who did not have a methylated MGMT promoter did not have such a benefit. [25] TMZ-based therapy is the standard of care for patients with GBM and resistance to this drug in GBM is modulated by MGMT. The MGMT promoter was found to be methylated in 45% of 206 assessable cases by Hegi et al. [25] Irrespective of treatment, MGMT promoter methylation was an independent favorable prognostic factor; among patients whose tumor contained a methylated MGMT promoter, survival benefit was observed in those treated with TMZ and radiotherapy. [25] Clinical studies have shown that elevated MGMT protein levels or lack of MGMT promoter methylation is associated with TMZ resistance in some GBM tumors. Kitange et al., also showed that MGMT expression is dynamically regulated in some non-methylated tumors and in these tumors, protracted dosing regimens may not be effective. [26]
Opportunities for improving esophageal cancer therapy include targeting epigenetic changes that occur with MGMT methylation. Such methylation may represent an early event in tumorigenesis, particularly in the setting of esophageal adenocarcinoma. [20] It has been well-established that esophageal adenocarcinoma can arise from Barrett’s esophagus. These changes may result in development of intestinal metaplasia with progression to intraepithelial neoplasia, and subsequently invasive adenocarcinoma. It has been demonstrated that hypermethylated CpG islands located in the promoter region of the MGMT exist at different steps in the progression to adenocarcinoma, suggesting a step-wise loss of DNA repair resulting in irreversible cellular damage. [16],[20] Use of an alkylating agent, such as TMZ, in the setting of ineffective DNA repair may result in sufficient DNA damage so as to result in apoptosis.
Much of the literature regarding the use of TMZ stems from the treatment of GBM. In this condition, it has been demonstrated that tumors lacking MGMT activity are more sensitive to the cytotoxic effects of TMZ than cells that express functional MGMT. In addition, tumors with high MGMT protein expression have demonstrated TMZ resistance. Reduced MGMT expression is seen in the setting of methylation of the MGMT promoter, which has been seen in 45% of cases. [25] In this disease, MGMT promoter methylation confers a survival benefit in patients treated with TMZ and radiotherapy presumably as a result of the cytotoxic effects of unrepaired O-6 methylguanine. [25] It can be postulated that a similar response to TMZ may be seen in esophageal cancer, another apoptosis-resistant cancer. [27]
In another investigation of the use of TMZ with esophageal cancer, Bruyere et al., [27] explored the in vitro and in vivo effect of TMZ on OE21 and OE33, esophageal cell lines. These studies demonstrated significant delays in OE21 xenograft tumor development in treated samples. Furthermore, there was improved survival in mice treated with TMZ compared with cisplatin.
Aside from the cytotoxicity, TMZ may also have a role in the inhibition of angiogenesis. In the Bruyere study, [27] xenografts demonstrated significantly decreased global tumor vascularization in TMZ treated mice. They also demonstrated down regulation of the proangiogenic chemokine chemokine (C-X-C motif) ligand (CXCL) and the angiogenic protein ID1, suggesting a VEGF independent mechanism of action. Combining TMZ with a standard esophageal cancer treatment may present a potentially new treatment option particularly in patients known to have MGMT methylation.
As a large number of studies are carried out to molecularly dissect esophageal adenocarcinoma versus SCC, we also should ensure that epigenetic changes are investigated. It is encouraging that certain subsets of esophageal tumors have MGMT methylation. It would be important to determine the relative concordance of other genetic aberrancies (such as HER2/neu amplification, MET amplification) with MGMT methylation.
Conclusion
Esophageal cancer is a difficult disease to treat and minimal personalized molecular markers exist. We present a unique case of an esophageal cancer patient with MGMT methylation who responded dramatically to TMZ. In light of this, a retrospective analysis was carried out on the frequency of MGMT methylation in squamous cell and adenocarcinoma of the esophagus. In vitro and in vivo modeling demonstrated the relative efficacy of TMZ in MGMT methylated cell lines. This study would then lead us to a MGMT methylation selected clinical trial for patients with esophageal cancer.
References
| 1. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69-90.  |
| 2. | Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: Defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 2006;24:2137-50.  |
| 3. | Gebski V, Burmeister B, Smithers BM, Foo K, Zalcberg J, Simes J, et al. Survival benefits from neoadjuvant chemoradiotherapy or chemotherapy in oesophageal carcinoma: A meta-analysis. Lancet Oncol 2007;8:226-34.  |
| 4. | Sjoquist KM, Burmeister BH, Smithers BM, Zalcberg JR, Simes RJ, Barbour A, et al. Survival after neoadjuvant chemotherapy or chemoradiotherapy for resectable oesophageal carcinoma: An updated meta-analysis. Lancet Oncol 2011;12:681-92.  |
| 5. | Hanawa M, Suzuki S, Dobashi Y, Yamane T, Kono K, Enomoto N, et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer 2006;118:1173-80.  |
| 6. | Karayiannakis AJ, Syrigos KN, Polychronidis A, Zbar A, Kouraklis G, Simopoulos C, et al. Circulating VEGF levels in the serum of gastric cancer patients: Correlation with pathological variables, patient survival, and tumor surgery. Ann Surg 2002;236:37-42.  |
| 7. | Wang KL, Wu TT, Choi IS, Wang H, Resetkova E, Correa AM, et al. Expression of epidermal growth factor receptor in esophageal and esophagogastric junction adenocarcinomas: Association with poor outcome. Cancer 2007;109:658-67.  |
| 8. | Gold PJ, Goldman B, Iqbal S, Leichman LP, Zhang W, Lenz HJ, et al. Cetuximab as second-line therapy in patients with metastatic esophageal adenocarcinoma: A phase II southwest oncology group study (S0415). J Thorac Oncol 2010;5:1472-6.  |
| 9. | Lorenzen S, Schuster T, Porschen R, Al-Batran SE, Hofheinz R, Thuss-Patience P, et al. Cetuximab plus cisplatin-5-fluorouracil versus cisplatin-5-fluorouracil alone in first-line metastatic squamous cell carcinoma of the esophagus: A randomized phase II study of the Arbeitsgemeinschaft Internistische Onkologie. Ann Oncol 2009;20:1667-73.  |
| 10. | Shah MA, Ramanathan RK, Ilson DH, Levnor A, D›Adamo D, O’Reilly E, et al. Multicenter phase II study of irinotecan, cisplatin, and bevacizumab in patients with metastatic gastric or gastroesophageal junction adenocarcinoma. J Clin Oncol 2006;24:5201-6.  |
| 11. | Spigel DR, Greco FA, Meluch AA, Lane CM, Farley C, Gray JR, et al. Phase I/II trial of preoperative oxaliplatin, docetaxel, and capecitabine with concurrent radiation therapy in localized carcinoma of the esophagus or gastroesophageal junction. J Clin Oncol 2010;28:2213-9.  |
| 12. | Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): A phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97.  |
| 13. | Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K, et al. MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 2011;29:4803-10.  |
| 14. | Hasina R, Mollberg N, Kawada I, Mutreja K, Kanade G, Yala S, et al. Critical role for the receptor tyrosine kinase EPHB4 in esophageal cancers. Cancer Res 2013;73:184-94.  |
| 15. | Baumann S, Keller G, Pühringer F, Napieralski R, Feith M, Langer R, et al. The prognostic impact of O6-Methylguanine-DNA Methyltransferase (MGMT) promotor hypermethylation in esophageal adenocarcinoma. Int J Cancer 2006;119:264-8.  |
| 16. | Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res 2001;61:3410-8.  |
| 17. | Schildhaus HU, Kröckel I, Lippert H, Malfertheiner P, Roessner A, Schneider-Stock R. Promoter hypermethylation of p16INK4a, E-cadherin, O6-MGMT, DAPK and FHIT in adenocarcinomas of the esophagus, esophagogastric junction and proximal stomach. Int J Oncol 2005;26:1493-500.  |
| 18. | Komine C, Watanabe T, Katayama Y, Yoshino A, Yokoyama T, Fukushima T. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol 2003;13:176-84.  |
| 19. | Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000;343:1350-4.  |
| 20. | Kuester D, El-Rifai W, Peng D, Ruemmele P, Kroeckel I, Peters B, et al. Silencing of MGMT expression by promoter hypermethylation in the metaplasia-dysplasia-carcinoma sequence of Barrett’s esophagus. Cancer Lett 2009;275:117-26.  |
| 21. | Fakhry C, D’souza G, Sugar E, Weber K, Goshu E, Minkoff H, et al. Relationship between prevalent oral and cervical human papillomavirus infections in human immunodeficiency virus-positive and-negative women. J Clin Microbiol 2006;44:4479-85.  |
| 22. | Friedman HS, McLendon RE, Kerby T, Dugan M, Bigner SH, Henry AJ, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol 1998;16:3851-7.  |
| 23. | Lai A, Tran A, Nghiemphu PL, Pope WB, Solis OE, Selch M, et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multiforme. J Clin Oncol 2011;29:142-8.  |
| 24. | Motomura K, Natsume A, Kishida Y, Higashi H, Kondo Y, Nakasu Y, et al. Benefits of interferon-β and temozolomide combination therapy for newly diagnosed primary glioblastoma with the unmethylated MGMT promoter: A multicenter study. Cancer 2011;117:1721-30.  |
| 25. | Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997-1003.  |
| 26. | Kitange GJ, Carlson BL, Schroeder MA, Grogan PT, Lamont JD, Decker PA, et al. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol 2009;11:281-91.  |
| 27. | Bruyère C, Lonez C, Duray A, Cludts S, Ruysschaert JM, Saussez S, et al. Considering temozolomide as a novel potential treatment for esophageal cancer. Cancer 2011;117:2004-16.  |
Authors
Rifat Hasina : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Mosmi Surati : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Ichiro Kawada : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Qudsia Arif : Department of Pathology, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
George B. Carey : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Rajani Kanteti : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Aliya N. Husain : Department of Pathology, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Mark K. Ferguson : Department of Surgery, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Everett E. Vokes : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Victoria M. Villaflor : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
Ravi Salgia : Department of Medicine, The University of Chicago, 5841 S Maryland Ave, MC 2115, Chicago, IL
